Selection of housekeeping genes for use in quantitative reverse transcription PCR assays on the murine cornea
- PMID: 20596249
- PMCID: PMC2893048
Selection of housekeeping genes for use in quantitative reverse transcription PCR assays on the murine cornea
Abstract
Purpose: To evaluate the suitability of common housekeeping genes (HKGs) for use in quantitative reverse transcription PCR (qRT-PCR) assays of the cornea in various murine disease models.
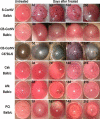
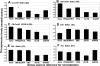
Methods: CORNEAL DISEASE MODELS STUDIED WERE: 1) corneal neovascularization (CorNV) induced by suture or chemical burn, 2) corneal infection with Candida albicans or Aspergillus fumigatus by intrastromal injection of live spores, and 3) perforating corneal injury (PCI) in Balb/c mice or C57BL/6 mice. Expression of 8 HKGs (glyceraldehyde-3-phosphate dehydrogenase [GAPDH], beta-actin [ACTB], lactate dehydrogenase A [LDHA], ribosomal protein L5 [RPL5], ubiquitin C [UBC], peptidylprolyl isomerase A [PPIA], TATA-box binding protein [TBP1], and hypoxanthine guanine phosphoribosyl transferase [HPRT1]) in the cornea were measured at various time points by microarray hybridization or qRT-PCR and the data analyzed using geNorm and NormFinder.
Results: Microarray results showed that under the CorNV condition the expression stability of the 8 HKGs decreased in order of PPIA>RPL5>HPRT1>ACTB>UBC>TBP1>GAPDH>LDHA. qRT-PCR analyses demonstrated that expression of none of the 8 HKGs remained stable under all conditions, while GAPDH and ACTB were among the least stably expressed markers under most conditions. Both geNorm and NormFinder analyses proposed best HKGs or HKG combinations that differ between the various models. NormFinder proposed PPIA as best HKG for three CorNV models and PCI model, as well as UBC for two fungal keratitis models. geNorm analysis demonstrated that a similar model in different mice strains or caused by different stimuli may require different HKGs or HKG pairs for the best normalization. Namely, geNorm proposed PPIA and HRPT1 and PPIA and RPL5 pairs for chemical burn-induced CorNV in Balb/c and C57BL/6 mice, respectively, while UBC and HPRT1 and UBC and LDHA were best for Candida and Aspergillus induced keratitis in Balb/c mice, respectively.
Conclusions: When qRT-PCR is designed for studies of gene expression in murine cornea, preselection of situation-specific reference genes is recommended. In the absence of knowledge about situation-specific HKGs, PPIA and UBC, either alone or in combination with HPRT1 or RPL5, can be employed.
Figures
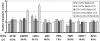


Similar articles
-
Validation of endogenous control genes for gene expression studies on human ocular surface epithelium.PLoS One. 2011;6(8):e22301. doi: 10.1371/journal.pone.0022301. Epub 2011 Aug 3. PLoS One. 2011. PMID: 21857920 Free PMC article.
-
Comparison of genome-wide gene expression in suture- and alkali burn-induced murine corneal neovascularization.Mol Vis. 2011;17:2386-99. Epub 2011 Sep 2. Mol Vis. 2011. PMID: 21921991 Free PMC article.
-
Identification of suitable reference genes for gene expression studies using quantitative polymerase chain reaction in lung cancer in vitro.Mol Med Rep. 2015 May;11(5):3767-73. doi: 10.3892/mmr.2015.3159. Epub 2015 Jan 8. Mol Med Rep. 2015. PMID: 25573171
-
Identification of valid reference housekeeping genes for gene expression analysis in tumor neovascularization studies.Clin Transl Oncol. 2013 Mar;15(3):211-8. doi: 10.1007/s12094-012-0904-1. Epub 2012 Jul 25. Clin Transl Oncol. 2013. PMID: 22855186
-
Expression and potential role of major inflammatory cytokines in experimental keratomycosis.Mol Vis. 2009 Jul 4;15:1303-11. Mol Vis. 2009. PMID: 19590756 Free PMC article.
Cited by
-
Reference genes for measuring mRNA expression.Theory Biosci. 2012 Dec;131(4):215-23. doi: 10.1007/s12064-012-0152-5. Epub 2012 May 17. Theory Biosci. 2012. PMID: 22588998 Review.
-
Differentiation of human embryonic stem cells into cells with corneal keratocyte phenotype.PLoS One. 2013;8(2):e56831. doi: 10.1371/journal.pone.0056831. Epub 2013 Feb 21. PLoS One. 2013. PMID: 23437251 Free PMC article.
-
Stability of Reference Genes for Messenger RNA Quantification by Real-Time PCR in Mouse Dextran Sodium Sulfate Experimental Colitis.PLoS One. 2016 May 31;11(5):e0156289. doi: 10.1371/journal.pone.0156289. eCollection 2016. PLoS One. 2016. PMID: 27244258 Free PMC article.
-
Animal Models of Aspergillosis.Comp Med. 2018 Apr 2;68(2):109-123. Comp Med. 2018. PMID: 29663936 Free PMC article. Review.
-
Identification and validation of suitable housekeeping genes for normalizing quantitative real-time PCR assays in injured peripheral nerves.PLoS One. 2014 Aug 21;9(8):e105601. doi: 10.1371/journal.pone.0105601. eCollection 2014. PLoS One. 2014. PMID: 25144298 Free PMC article.
References
-
- Thellin O, Zorzi W, Lakaye B, De Borman B, Coumans B, Hennen G, Grisar T, Igout A, Heinen E. Housekeeping genes as internal standards: use and limits. J Biotechnol. 1999;75:291–5. - PubMed
-
- Barber RD, Harmer DW, Coleman RA, Clark BJ. GAPDH as a housekeeping gene: analysis of GAPDH mRNA expression in a panel of 72 human tissues. Physiol Genomics. 2005;21:389–95. - PubMed
-
- Suzuki T, Higgins PJ, Crawford DR. Control selection for RNA quantitation. Biotechniques. 2000;29:332–7. - PubMed
-
- Selvey S, Thompson EW, Matthaei K, Lea RA, Irving MG, Griffiths LR. Beta-actin–an unsuitable internal control for RT-PCR. Mol Cell Probes. 2001;15:307–11. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous
