Identification of broad-based HIV-1 protease inhibitors from combinatorial libraries
- PMID: 20507280
- PMCID: PMC3084599
- DOI: 10.1042/BJ20091645
Identification of broad-based HIV-1 protease inhibitors from combinatorial libraries
Abstract
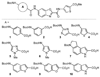
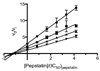
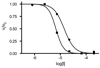
Clinically approved inhibitors of the HIV-1 protease function via a competitive mechanism. A particular vulnerability of competitive inhibitors is their sensitivity to increases in substrate concentration, as may occur during virion assembly, budding and processing into a mature infectious viral particle. Advances in chemical synthesis have led to the development of new high-diversity chemical libraries using rapid in-solution syntheses. These libraries have been shown previously to be effective at disrupting protein-protein and protein-nucleic acid interfaces. We have screened 44000 compounds from such a library to identify inhibitors of the HIV-1 protease. One compound was identified that inhibits wild-type protease, as well as a drug-resistant protease with six mutations. Moreover, analysis of this compound suggests an allosteric non-competitive mechanism of inhibition and may represent a starting point for an additional strategy for anti-retroviral therapy.
Figures
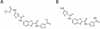


Similar articles
-
Design of mutation-resistant HIV protease inhibitors with the substrate envelope hypothesis.Chem Biol Drug Des. 2007 May;69(5):298-313. doi: 10.1111/j.1747-0285.2007.00514.x. Chem Biol Drug Des. 2007. PMID: 17539822
-
Beta-lactam compounds as apparently uncompetitive inhibitors of HIV-1 protease.Bioorg Med Chem Lett. 2005 Jun 15;15(12):3086-90. doi: 10.1016/j.bmcl.2005.04.020. Bioorg Med Chem Lett. 2005. PMID: 15893929
-
Resilience to resistance of HIV-1 protease inhibitors: profile of darunavir.AIDS Rev. 2008 Jul-Sep;10(3):131-42. AIDS Rev. 2008. PMID: 18820715 Free PMC article. Review.
-
Combinatorial diversification of indinavir: in vivo mixture dosing of an HIV protease inhibitor library.Bioorg Med Chem Lett. 2000 Jul 17;10(14):1527-30. doi: 10.1016/s0960-894x(00)00276-6. Bioorg Med Chem Lett. 2000. PMID: 10915042
-
Structural and thermodynamic basis of resistance to HIV-1 protease inhibition: implications for inhibitor design.Curr Drug Targets Infect Disord. 2003 Dec;3(4):311-28. doi: 10.2174/1568005033481051. Curr Drug Targets Infect Disord. 2003. PMID: 14754432 Review.
Cited by
-
Discovery libraries targeting the major enzyme classes: the serine hydrolases.Bioorg Med Chem Lett. 2014 Aug 15;24(16):3807-13. doi: 10.1016/j.bmcl.2014.06.063. Epub 2014 Jun 27. Bioorg Med Chem Lett. 2014. PMID: 25037918 Free PMC article.
-
Design, synthesis, and validation of a β-turn mimetic library targeting protein-protein and peptide-receptor interactions.J Am Chem Soc. 2011 Jul 6;133(26):10184-94. doi: 10.1021/ja201878v. Epub 2011 Jun 15. J Am Chem Soc. 2011. PMID: 21609016 Free PMC article.
-
Novel approaches in anti-arenaviral drug development.Virology. 2011 Mar 15;411(2):163-9. doi: 10.1016/j.virol.2010.11.022. Epub 2010 Dec 22. Virology. 2011. PMID: 21183197 Free PMC article. Review.
-
Flexible catalytic site conformations implicated in modulation of HIV-1 protease autoprocessing reactions.Retrovirology. 2011 Oct 10;8:79. doi: 10.1186/1742-4690-8-79. Retrovirology. 2011. PMID: 21985091 Free PMC article.
-
High-throughput screening for the identification of small-molecule inhibitors of the flaviviral protease.Antiviral Res. 2016 Oct;134:6-16. doi: 10.1016/j.antiviral.2016.08.014. Epub 2016 Aug 15. Antiviral Res. 2016. PMID: 27539384 Free PMC article.
References
-
- UNAIDS. Joint United Nations Programme on HIV/AIDS. Geneva: 2008. 2008 Report on the global AIDS epidemic.
-
- Bennett DE, Camacho RJ, Otelea D, Kuritzkes DR, Fleury H, Kiuchi M, Heneine W, Kantor R, Jordan MR, Schapiro JM, Vandamme AM, Sandstrom P, Boucher CA, van de Vijver D, Rhee SY, Liu TF, Pillay D, Shafer RW. Drug resistance mutations for surveillance of transmitted HIV-1 drug-resistance: 2009 update. PLoS ONE. 2009;4:e4724. - PMC - PubMed
-
- Copeland RA. Evaluation of enzyme inhibitors in drug discovery : a guide for medicinal chemists and pharmacologists. Hoboken, N.J.: Wiley-Interscience; 2005. - PubMed
-
- Bowman MJ, Byrne S, Chmielewski J. Switching between allosteric and dimerization inhibition of HIV-1 protease. Chem. Biol. 2005;12:439–444. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AI040882/AI/NIAID NIH HHS/United States
- GM48870/GM/NIGMS NIH HHS/United States
- P01 GM048870-08/GM/NIGMS NIH HHS/United States
- P01 GM083658/GM/NIGMS NIH HHS/United States
- R01 AI040882-03/AI/NIAID NIH HHS/United States
- P01 GM083658-01/GM/NIGMS NIH HHS/United States
- P01 CA078045/CA/NCI NIH HHS/United States
- P30 AI036214/AI/NIAID NIH HHS/United States
- T32 AI007354-09/AI/NIAID NIH HHS/United States
- P01 GM048870/GM/NIGMS NIH HHS/United States
- GM083658/GM/NIGMS NIH HHS/United States
- T32 AI007354/AI/NIAID NIH HHS/United States
- P30 AI036214-13S1/AI/NIAID NIH HHS/United States
- P01 CA078045-02/CA/NCI NIH HHS/United States
- 3 P30 AI036214-13S1/AI/NIAID NIH HHS/United States
- 5T32AI007354/AI/NIAID NIH HHS/United States
- T32 NS041219/NS/NINDS NIH HHS/United States
- AI40882/AI/NIAID NIH HHS/United States
- 5T32NSO412119/PHS HHS/United States
- CA78045/CA/NCI NIH HHS/United States
- T32 NS041219-01/NS/NINDS NIH HHS/United States
LinkOut - more resources
Full Text Sources

