Sti1 regulation of Hsp70 and Hsp90 is critical for curing of Saccharomyces cerevisiae [PSI+] prions by Hsp104
- PMID: 20479121
- PMCID: PMC2897543
- DOI: 10.1128/MCB.01292-09
Sti1 regulation of Hsp70 and Hsp90 is critical for curing of Saccharomyces cerevisiae [PSI+] prions by Hsp104
Abstract
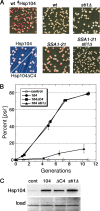
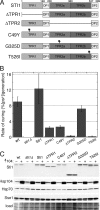
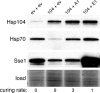
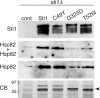
Although propagation of Saccharomyces cerevisiae prions requires Hsp104 protein disaggregating activity, overproducing Hsp104 "cures" cells of [PSI(+)] prions. Earlier evidence suggests that the Hsp70 mutant Ssa1-21 impairs [PSI(+)] by a related mechanism. Here, we confirm this link by finding that deletion of STI1 both suppresses Ssa1-21 impairment of [PSI(+)] and blocks Hsp104 curing of [PSI(+)]. Hsp104's tetratricopeptide repeat (TPR) interaction motif was dispensable for curing; however, cells expressing Sti1 defective in Hsp70 or Hsp90 interaction cured less efficiently, and the Hsp90 inhibitor radicicol abolished curing, implying that Sti1 acts in curing through Hsp70 and Hsp90 interactions. Accordingly, strains lacking constitutive or inducible Hsp90 isoforms cured at reduced rates. We confirm an earlier finding that elevating free ubiquitin levels enhances curing, but it did not overcome inhibition of curing caused by Hsp90 defects, suggesting that Hsp90 machinery is important for the contribution of ubiquitin to curing. We also find curing associated with cell division. Our findings point to crucial roles of Hsp70, Sti1, and Hsp90 for efficient curing by overexpressed Hsp104 and provide evidence supporting the earlier suggestion that destruction of prions by protein disaggregation does not adequately explain the curing.
Figures

Similar articles
-
Propagation of Saccharomyces cerevisiae [PSI+] prion is impaired by factors that regulate Hsp70 substrate binding.Mol Cell Biol. 2004 May;24(9):3928-37. doi: 10.1128/MCB.24.9.3928-3937.2004. Mol Cell Biol. 2004. PMID: 15082786 Free PMC article.
-
N-terminal domain of yeast Hsp104 chaperone is dispensable for thermotolerance and prion propagation but necessary for curing prions by Hsp104 overexpression.Genetics. 2006 Jun;173(2):611-20. doi: 10.1534/genetics.106.056820. Epub 2006 Apr 2. Genetics. 2006. PMID: 16582428 Free PMC article.
-
Antagonistic interactions between yeast chaperones Hsp104 and Hsp70 in prion curing.Mol Cell Biol. 1999 Feb;19(2):1325-33. doi: 10.1128/MCB.19.2.1325. Mol Cell Biol. 1999. PMID: 9891066 Free PMC article.
-
The life of [PSI].Curr Genet. 2018 Feb;64(1):1-8. doi: 10.1007/s00294-017-0714-7. Epub 2017 Jun 26. Curr Genet. 2018. PMID: 28653109 Free PMC article. Review.
-
Remembering the Past: A New Form of Protein-Based Inheritance.Cell. 2016 Oct 6;167(2):302-303. doi: 10.1016/j.cell.2016.09.036. Cell. 2016. PMID: 27716500 Review.
Cited by
-
Prokaryotic chaperones support yeast prions and thermotolerance and define disaggregation machinery interactions.Genetics. 2012 Sep;192(1):185-93. doi: 10.1534/genetics.112.142307. Epub 2012 Jun 25. Genetics. 2012. PMID: 22732191 Free PMC article.
-
Yeast and Fungal Prions.Cold Spring Harb Perspect Biol. 2016 Sep 1;8(9):a023531. doi: 10.1101/cshperspect.a023531. Cold Spring Harb Perspect Biol. 2016. PMID: 27481532 Free PMC article. Review.
-
The hop-like stress-induced protein 1 cochaperone is a novel cell-intrinsic restriction factor for mitochondrial tombusvirus replication.J Virol. 2014 Aug;88(16):9361-78. doi: 10.1128/JVI.00561-14. Epub 2014 Jun 11. J Virol. 2014. PMID: 24920799 Free PMC article.
-
Increased levels of Stress-inducible phosphoprotein-1 accelerates amyloid-β deposition in a mouse model of Alzheimer's disease.Acta Neuropathol Commun. 2020 Aug 21;8(1):143. doi: 10.1186/s40478-020-01013-5. Acta Neuropathol Commun. 2020. PMID: 32825842 Free PMC article.
-
Overexpression of Hsp104 by Causing Dissolution of the Prion Seeds Cures the Yeast [PSI+] Prion.Int J Mol Sci. 2023 Jun 29;24(13):10833. doi: 10.3390/ijms241310833. Int J Mol Sci. 2023. PMID: 37446010 Free PMC article.
References
-
- Acebron, S. P., I. Martin, U. del Castillo, F. Moro, and A. Muga. 2009. DnaK-mediated association of ClpB to protein aggregates. A bichaperone network at the aggregate surface. FEBS Lett. 583:2991-2996. - PubMed
-
- Allen, K. D., T. A. Chernova, E. P. Tennant, K. D. Wilkinson, and Y. O. Chernoff. 2007. Effects of ubiquitin system alterations on the formation and loss of a yeast prion. J. Biol. Chem. 282:3004-3013. - PubMed
-
- Allen, K. D., R. D. Wegrzyn, T. A. Chernova, S. Muller, G. P. Newnam, P. A. Winslett, K. B. Wittich, K. D. Wilkinson, and Y. O. Chernoff. 2005. Hsp70 chaperones as modulators of prion life cycle: novel effects of Ssa and Ssb on the Saccharomyces cerevisiae prion [PSI+]. Genetics 169:1227-1242. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
