MicroRNAs, macrocontrol: regulation of miRNA processing
- PMID: 20423980
- PMCID: PMC2874160
- DOI: 10.1261/rna.1804410
MicroRNAs, macrocontrol: regulation of miRNA processing
Abstract
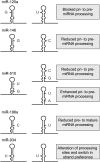
MicroRNAs (miRNAs) are a set of small, non-protein-coding RNAs that regulate gene expression at the post-transcriptional level. Maturation of miRNAs comprises several regulated steps resulting in approximately 22-nucleotide single-stranded mature miRNAs. Regulation of miRNA expression can occur both at the transcriptional level and at the post-transcriptional level during miRNA processing. Recent studies have elucidated specific aspects of the well-regulated nature of miRNA processing involving various regulatory proteins, editing of miRNA transcripts, and cellular location. In addition, single nucleotide polymorphisms in miRNA genes can also affect the processing efficiency of primary miRNA transcripts. In this review we present an overview of the currently known regulatory pathways of miRNA processing and provide a basis to understand how aberrant miRNA processing may arise and may be involved in pathophysiological conditions such as cancer.
Figures
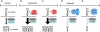

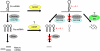
Similar articles
-
Improved annotation of C. elegans microRNAs by deep sequencing reveals structures associated with processing by Drosha and Dicer.RNA. 2011 Apr;17(4):563-77. doi: 10.1261/rna.2432311. Epub 2011 Feb 9. RNA. 2011. PMID: 21307183 Free PMC article.
-
Processing of primary microRNAs by the Microprocessor complex.Nature. 2004 Nov 11;432(7014):231-5. doi: 10.1038/nature03049. Epub 2004 Nov 7. Nature. 2004. PMID: 15531879
-
Autoregulation of microRNA biogenesis by let-7 and Argonaute.Nature. 2012 Jun 28;486(7404):541-4. doi: 10.1038/nature11134. Nature. 2012. PMID: 22722835 Free PMC article.
-
Drosha in primary microRNA processing.Cold Spring Harb Symp Quant Biol. 2006;71:51-7. doi: 10.1101/sqb.2006.71.041. Cold Spring Harb Symp Quant Biol. 2006. PMID: 17381280 Review.
-
Smad-mediated miRNA processing: a critical role for a conserved RNA sequence.RNA Biol. 2011 Jan-Feb;8(1):71-6. doi: 10.4161/rna.8.1.14299. Epub 2011 Jan 1. RNA Biol. 2011. PMID: 21289485 Free PMC article. Review.
Cited by
-
Virus-encoded microRNAs: an overview and a look to the future.PLoS Pathog. 2012 Dec;8(12):e1003018. doi: 10.1371/journal.ppat.1003018. Epub 2012 Dec 20. PLoS Pathog. 2012. PMID: 23308061 Free PMC article. Review.
-
Arm Selection Preference of MicroRNA-193a Varies in Breast Cancer.Sci Rep. 2016 Jun 16;6:28176. doi: 10.1038/srep28176. Sci Rep. 2016. PMID: 27307030 Free PMC article.
-
DICER governs characteristics of glioma stem cells and the resulting tumors in xenograft mouse models of glioblastoma.Oncotarget. 2016 Aug 30;7(35):56431-56446. doi: 10.18632/oncotarget.10570. Oncotarget. 2016. PMID: 27421140 Free PMC article.
-
Emerging microRNA biomarkers for colorectal cancer diagnosis and prognosis.Open Biol. 2019 Jan 31;9(1):180212. doi: 10.1098/rsob.180212. Open Biol. 2019. PMID: 30958116 Free PMC article. Review.
-
Comprehensive identification of microRNA arm selection preference in lung cancer: miR-324-5p and -3p serve oncogenic functions in lung cancer.Oncol Lett. 2018 Jun;15(6):9818-9826. doi: 10.3892/ol.2018.8557. Epub 2018 Apr 24. Oncol Lett. 2018. PMID: 29844840 Free PMC article.
References
-
- Amariglio N, Rechavi G 2007. A-to-I RNA editing: A new regulatory mechanism of global gene expression. Blood Cells Mol 39: 151–155 - PubMed
-
- Bartel DP 2004. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 116: 281–297 - PubMed
-
- Calin GA, Ferracin M, Cimmino A, Di Leva G, Shimizu M, Wojcik SE, Iorio MV, Visone R, Sever NI, Fabbri M, et al. 2005. A microRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med 353: 1793–1801 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
