FGFR1 amplification drives endocrine therapy resistance and is a therapeutic target in breast cancer
- PMID: 20179196
- PMCID: PMC2832818
- DOI: 10.1158/0008-5472.CAN-09-3746
FGFR1 amplification drives endocrine therapy resistance and is a therapeutic target in breast cancer
Abstract
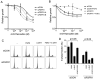
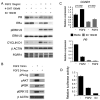
Amplification of fibroblast growth factor receptor 1 (FGFR1) occurs in approximately 10% of breast cancers and is associated with poor prognosis. However, it is uncertain whether overexpression of FGFR1 is causally linked to the poor prognosis of amplified cancers. Here, we show that FGFR1 overexpression is robustly associated with FGFR1 amplification in two independent series of breast cancers. Breast cancer cell lines with FGFR1 overexpression and amplification show enhanced ligand-dependent signaling, with increased activation of the mitogen-activated protein kinase and phosphoinositide 3-kinase-AKT signaling pathways in response to FGF2, but also show basal ligand-independent signaling, and are dependent on FGFR signaling for anchorage-independent growth. FGFR1-amplified cell lines show resistance to 4-hydroxytamoxifen, which is reversed by small interfering RNA silencing of FGFR1, suggesting that FGFR1 overexpression also promotes endocrine therapy resistance. FGFR1 signaling suppresses progesterone receptor (PR) expression in vitro, and likewise, amplified cancers are frequently PR negative, identifying a potential biomarker for FGFR1 activity. Furthermore, we show that amplified cancers have a high proliferative rate assessed by Ki67 staining and that FGFR1 amplification is found in 16% to 27% of luminal B-type breast cancers. Our data suggest that amplification and overexpression of FGFR1 may be a major contributor to poor prognosis in luminal-type breast cancers, driving anchorage-independent proliferation and endocrine therapy resistance.
Figures


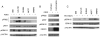

Comment in
-
Genetic alterations of FGF receptors: an emerging field in clinical cancer diagnostics and therapeutics.Expert Rev Anticancer Ther. 2010 Sep;10(9):1375-9. doi: 10.1586/era.10.128. Expert Rev Anticancer Ther. 2010. PMID: 20836672
Similar articles
-
FGFR1 amplification or overexpression and hormonal resistance in luminal breast cancer: rationale for a triple blockade of ER, CDK4/6, and FGFR1.Breast Cancer Res. 2021 Feb 12;23(1):21. doi: 10.1186/s13058-021-01398-8. Breast Cancer Res. 2021. PMID: 33579347 Free PMC article.
-
Association of FGFR1 with ERα Maintains Ligand-Independent ER Transcription and Mediates Resistance to Estrogen Deprivation in ER+ Breast Cancer.Clin Cancer Res. 2017 Oct 15;23(20):6138-6150. doi: 10.1158/1078-0432.CCR-17-1232. Epub 2017 Jul 27. Clin Cancer Res. 2017. PMID: 28751448 Free PMC article.
-
Overcoming endocrine resistance due to reduced PTEN levels in estrogen receptor-positive breast cancer by co-targeting mammalian target of rapamycin, protein kinase B, or mitogen-activated protein kinase kinase.Breast Cancer Res. 2014 Sep 11;16(5):430. doi: 10.1186/s13058-014-0430-x. Breast Cancer Res. 2014. PMID: 25212826 Free PMC article.
-
Fibroblast growth factor receptor signaling in estrogen receptor-positive breast cancer: mechanisms and role in endocrine resistance.Front Oncol. 2024 Jul 8;14:1406951. doi: 10.3389/fonc.2024.1406951. eCollection 2024. Front Oncol. 2024. PMID: 39040443 Free PMC article. Review.
-
Acquired endocrine resistance in breast cancer: implications for tumour metastasis.Front Biosci (Landmark Ed). 2011 Jan 1;16(3):838-48. doi: 10.2741/3723. Front Biosci (Landmark Ed). 2011. PMID: 21196206 Review.
Cited by
-
The genomic landscape of breast cancer as a therapeutic roadmap.Cancer Discov. 2013 Jan;3(1):27-34. doi: 10.1158/2159-8290.CD-12-0462. Cancer Discov. 2013. PMID: 23319768 Free PMC article.
-
Clinical Implementation of Novel Targeted Therapeutics in Advanced Breast Cancer.J Cell Biochem. 2016 Nov;117(11):2454-63. doi: 10.1002/jcb.25590. Epub 2016 Jun 3. J Cell Biochem. 2016. PMID: 27146558 Free PMC article. Review.
-
Luminal-B breast cancer and novel therapeutic targets.Breast Cancer Res. 2011;13(6):221. doi: 10.1186/bcr2904. Epub 2011 Nov 30. Breast Cancer Res. 2011. PMID: 22217398 Free PMC article. Review.
-
FGFR signaling maintains a drug persistent cell population following epithelial-mesenchymal transition.Oncotarget. 2016 Dec 13;7(50):83424-83436. doi: 10.18632/oncotarget.13117. Oncotarget. 2016. PMID: 27825137 Free PMC article.
-
The transition model of RTK activation: A quantitative framework for understanding RTK signaling and RTK modulator activity.Cytokine Growth Factor Rev. 2019 Oct;49:23-31. doi: 10.1016/j.cytogfr.2019.10.004. Epub 2019 Nov 1. Cytokine Growth Factor Rev. 2019. PMID: 31711797 Free PMC article. Review.
References
-
- Perou CM, Sorlie T, Eisen MB, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–52. - PubMed
-
- Loi S, Haibe-Kains B, Desmedt C, et al. Definition of clinically distinct molecular subtypes in estrogen receptor-positive breast carcinomas through genomic grade. J Clin Oncol. 2007;25:1239–46. - PubMed
-
- Paik S, Shak S, Tang G, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351:2817–26. - PubMed
-
- Courjal F, Cuny M, Simony-Lafontaine J, et al. Mapping of DNA amplifications at 15 chromosomal localizations in 1875 breast tumors: definition of phenotypic groups. Cancer research. 1997;57:4360–7. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

