Kaposi sarcoma-associated herpesvirus latency-associated nuclear antigen inhibits interferon (IFN) beta expression by competing with IFN regulatory factor-3 for binding to IFNB promoter
- PMID: 20048166
- PMCID: PMC2844170
- DOI: 10.1074/jbc.M109.018838
Kaposi sarcoma-associated herpesvirus latency-associated nuclear antigen inhibits interferon (IFN) beta expression by competing with IFN regulatory factor-3 for binding to IFNB promoter
Abstract
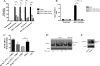
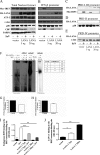
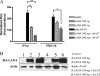
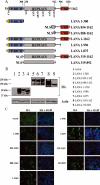
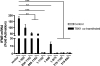
Host cells respond to viral infections by synthesizing and producing antiviral molecules such as type I interferons (IFN). The Kaposi sarcoma-associated herpesvirus (KSHV) encodes multiple proteins expressed during the lytic replication cycle that alter the antiviral response of the host. Considering that in Kaposi sarcoma lesions and primary effusion lymphoma cells KSHV is latent in the vast majority of cells, we were interested in determining whether latently expressed viral proteins have the ability to modulate IFN synthesis. The latency-associated nuclear antigen (LANA-1) is a large nuclear protein that plays a role in the establishment and maintenance of latent KSHV episome in the nucleus of infected cells. LANA-1 is also described to modulate the cellular transcription. Here, we report that LANA-1 inhibits IFN-beta transcription and synthesis by competing with the binding of interferon regulatory factor-3 (IRF3) to the IFNB promoter. Using mutants of LANA-1, we have identified the central acidic repeated region as the domain essential for interfering with the binding of IRF3 to the positive regulatory domains I-III of the IFNB promoter. In addition, the nuclear localization of LANA-1 proved essential for IFN-beta inhibition. Thus, LANA-1 interferes with the formation of IFN-beta enhanceosome by competing with the fixation of IRF3 and by inhibiting the expression of the CREB-binding protein. The ability of LANA-1 to inhibit IFNB gene expression highlights a new role for this protein in cellular gene modulation and immune evasion strategies.
Figures


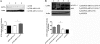
Similar articles
-
Activated Nrf2 Interacts with Kaposi's Sarcoma-Associated Herpesvirus Latency Protein LANA-1 and Host Protein KAP1 To Mediate Global Lytic Gene Repression.J Virol. 2015 Aug;89(15):7874-92. doi: 10.1128/JVI.00895-15. Epub 2015 May 20. J Virol. 2015. PMID: 25995248 Free PMC article.
-
Kaposi's Sarcoma-Associated Herpesvirus Latency-Associated Nuclear Antigen Inhibits Major Histocompatibility Complex Class II Expression by Disrupting Enhanceosome Assembly through Binding with the Regulatory Factor X Complex.J Virol. 2015 May;89(10):5536-56. doi: 10.1128/JVI.03713-14. Epub 2015 Mar 4. J Virol. 2015. PMID: 25740990 Free PMC article.
-
Site-specific association with host and viral chromatin by Kaposi's sarcoma-associated herpesvirus LANA and its reversal during lytic reactivation.J Virol. 2014 Jun;88(12):6762-77. doi: 10.1128/JVI.00268-14. Epub 2014 Apr 2. J Virol. 2014. PMID: 24696474 Free PMC article.
-
Kaposi sarcoma-associated herpesvirus latency-associated nuclear antigen: more than a key mediator of viral persistence.Curr Opin Virol. 2023 Aug;61:101336. doi: 10.1016/j.coviro.2023.101336. Epub 2023 Jun 16. Curr Opin Virol. 2023. PMID: 37331160 Review.
-
KSHV Genome Replication and Maintenance in Latency.Adv Exp Med Biol. 2018;1045:299-320. doi: 10.1007/978-981-10-7230-7_14. Adv Exp Med Biol. 2018. PMID: 29896673 Review.
Cited by
-
Kaposi's Sarcoma-Associated Herpesvirus Reduces Cellular Myeloid Differentiation Primary-Response Gene 88 (MyD88) Expression via Modulation of Its RNA.J Virol. 2015 Oct 14;90(1):180-8. doi: 10.1128/JVI.02342-15. Print 2016 Jan 1. J Virol. 2015. PMID: 26468534 Free PMC article.
-
Cytoplasmic isoforms of Kaposi sarcoma herpesvirus LANA recruit and antagonize the innate immune DNA sensor cGAS.Proc Natl Acad Sci U S A. 2016 Feb 23;113(8):E1034-43. doi: 10.1073/pnas.1516812113. Epub 2016 Jan 25. Proc Natl Acad Sci U S A. 2016. PMID: 26811480 Free PMC article.
-
Identification of host-chromosome binding sites and candidate gene targets for Kaposi's sarcoma-associated herpesvirus LANA.J Virol. 2012 May;86(10):5752-62. doi: 10.1128/JVI.07216-11. Epub 2012 Mar 14. J Virol. 2012. PMID: 22419807 Free PMC article.
-
Role of IRF4 in IFN-stimulated gene induction and maintenance of Kaposi sarcoma-associated herpesvirus latency in primary effusion lymphoma cells.J Immunol. 2013 Aug 1;191(3):1476-85. doi: 10.4049/jimmunol.1202514. Epub 2013 Jun 26. J Immunol. 2013. PMID: 23804715 Free PMC article.
-
Of Keeping and Tipping the Balance: Host Regulation and Viral Modulation of IRF3-Dependent IFNB1 Expression.Viruses. 2020 Jul 7;12(7):733. doi: 10.3390/v12070733. Viruses. 2020. PMID: 32645843 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

