Regulation of KSHV lytic switch protein expression by a virus-encoded microRNA: an evolutionary adaptation that fine-tunes lytic reactivation
- PMID: 20006845
- PMCID: PMC2822622
- DOI: 10.1016/j.chom.2009.11.008
Regulation of KSHV lytic switch protein expression by a virus-encoded microRNA: an evolutionary adaptation that fine-tunes lytic reactivation
Abstract
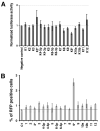
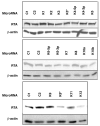
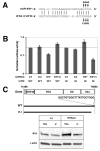
Herpesviruses encode numerous microRNAs (miRNAs), most of whose functions are unknown. The Kaposi's sarcoma-associated herpesvirus (KSHV) encodes 17 known miRNAs as part of its latency program, suggesting that these RNAs might function to regulate the latent state. Here we show that one of these KSHV miRNAs, miRK9( *), targets a sequence in the 3' untranslated region (UTR) of the mRNA encoding the major lytic switch protein (RTA), which controls viral reactivation from latency. Ectopic expression of miRK9( *) impairs RTA synthesis, while its specific antagonism in latently infected cells enhances spontaneous lytic reactivation frequency by 2- to 3-fold. Mutation of the recognition sequence in the RTA 3'UTR abolishes RTA downregulation by miRK9( *). We propose that miRNA targeting of RTA, while not the primary regulator of the lytic switch, functions like a safety mechanism on the trigger of lytic reactivation, preventing stochastic variations in basal RTA transcription from activating inappropriate entry into the lytic cycle.
Figures

Similar articles
-
Inhibition of Kaposi's sarcoma-associated herpesvirus lytic replication by HIV-1 Nef and cellular microRNA hsa-miR-1258.J Virol. 2014 May;88(9):4987-5000. doi: 10.1128/JVI.00025-14. Epub 2014 Feb 19. J Virol. 2014. PMID: 24554664 Free PMC article.
-
Genome-Wide Identification of Direct RTA Targets Reveals Key Host Factors for Kaposi's Sarcoma-Associated Herpesvirus Lytic Reactivation.J Virol. 2019 Feb 19;93(5):e01978-18. doi: 10.1128/JVI.01978-18. Print 2019 Mar 1. J Virol. 2019. PMID: 30541837 Free PMC article.
-
CCAAT/enhancer-binding protein-alpha is induced during the early stages of Kaposi's sarcoma-associated herpesvirus (KSHV) lytic cycle reactivation and together with the KSHV replication and transcription activator (RTA) cooperatively stimulates the viral RTA, MTA, and PAN promoters.J Virol. 2003 Sep;77(17):9590-612. doi: 10.1128/jvi.77.17.9590-9612.2003. J Virol. 2003. PMID: 12915572 Free PMC article.
-
KSHV miRNAs decrease expression of lytic genes in latently infected PEL and endothelial cells by targeting host transcription factors.Viruses. 2014 Oct 23;6(10):4005-23. doi: 10.3390/v6104005. Viruses. 2014. PMID: 25341664 Free PMC article. Review.
-
The regulation of KSHV lytic reactivation by viral and cellular factors.Curr Opin Virol. 2022 Feb;52:39-47. doi: 10.1016/j.coviro.2021.11.004. Epub 2021 Dec 3. Curr Opin Virol. 2022. PMID: 34872030 Free PMC article. Review.
Cited by
-
Kaposi's sarcoma-associated herpesvirus microRNAs repress breakpoint cluster region protein expression, enhance Rac1 activity, and increase in vitro angiogenesis.J Virol. 2015 Apr;89(8):4249-61. doi: 10.1128/JVI.03687-14. Epub 2015 Jan 28. J Virol. 2015. PMID: 25631082 Free PMC article.
-
A Kaposi's sarcoma-associated herpesvirus microRNA and its variants target the transforming growth factor β pathway to promote cell survival.J Virol. 2012 Nov;86(21):11698-711. doi: 10.1128/JVI.06855-11. Epub 2012 Aug 22. J Virol. 2012. PMID: 22915806 Free PMC article.
-
Herpes Simplex Virus 1 Deregulation of Host MicroRNAs.Noncoding RNA. 2018 Nov 23;4(4):36. doi: 10.3390/ncrna4040036. Noncoding RNA. 2018. PMID: 30477082 Free PMC article. Review.
-
Viruses, microRNAs, and host interactions.Annu Rev Microbiol. 2010;64:123-41. doi: 10.1146/annurev.micro.112408.134243. Annu Rev Microbiol. 2010. PMID: 20477536 Free PMC article. Review.
-
MicroRNA Regulation of Human Herpesvirus Latency.Viruses. 2022 Jun 2;14(6):1215. doi: 10.3390/v14061215. Viruses. 2022. PMID: 35746686 Free PMC article. Review.
References
-
- Blake WJ, Balazsi G, Kohanski MA, Isaacs FJ, Murphy KF, Kuang Y, Cantor CR, Walt DR, Collins JJ. Phenotypic consequences of promoter-mediated transcriptional noise. Molecular cell. 2006;24:853–865. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

