KSHV-induced notch components render endothelial and mural cell characteristics and cell survival
- PMID: 19965636
- PMCID: PMC2815507
- DOI: 10.1182/blood-2009-08-236745
KSHV-induced notch components render endothelial and mural cell characteristics and cell survival
Abstract
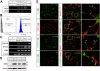
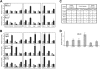
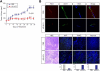
Kaposi sarcoma-associated herpesvirus (KSHV) infection is essential to the development of Kaposi sarcoma (KS). Notch signaling is also known to play a pivotal role in KS cell survival and lytic phase entrance of KSHV. In the current study, we sought to determine whether KSHV regulates Notch components. KSHV-infected lymphatic endothelial cells showed induction of receptors Notch3 and Notch4, Notch ligands Dll4 and Jagged1, and activated Notch receptors in contrast to uninfected lymphatic endothelial cells. In addition, KSHV induced the expression of endothelial precursor cell marker (CD133) and mural cell markers (calponin, desmin, and smooth muscle alpha actin), suggesting dedifferentiation and trans-differentiation. Overexpression of latency proteins (LANA, vFLIP) and lytic phase proteins (RTA, vGPCR, viral interleukin-6) further supported the direct regulatory capacity of KSHV viral proteins to induce Notch receptors (Notch2, Notch3), ligands (Dll1, Dll4, Jagged1), downstream targets (Hey, Hes), and endothelial precursor CD133. Targeting Notch pathway with gamma-secretase inhibitor and a decoy protein in the form of soluble Dll4 inhibited growth of KSHV-transformed endothelial cell line. Soluble Dll4 was also highly active in vivo against KS tumor xenograft. It inhibited tumor cell growth, induced tumor cell death, and reduced vessel perfusion. Soluble Dll4 is thus a candidate for clinical investigation.
Figures



Similar articles
-
KSHV manipulates Notch signaling by DLL4 and JAG1 to alter cell cycle genes in lymphatic endothelia.PLoS Pathog. 2009 Oct;5(10):e1000616. doi: 10.1371/journal.ppat.1000616. Epub 2009 Oct 9. PLoS Pathog. 2009. PMID: 19816565 Free PMC article.
-
Endothelial Jagged1 antagonizes Dll4 regulation of endothelial branching and promotes vascular maturation downstream of Dll4/Notch1.Arterioscler Thromb Vasc Biol. 2015 May;35(5):1134-46. doi: 10.1161/ATVBAHA.114.304741. Epub 2015 Mar 12. Arterioscler Thromb Vasc Biol. 2015. PMID: 25767274
-
NOTCH3 expression is induced in mural cells through an autoregulatory loop that requires endothelial-expressed JAGGED1.Circ Res. 2009 Feb 27;104(4):466-75. doi: 10.1161/CIRCRESAHA.108.184846. Epub 2009 Jan 15. Circ Res. 2009. PMID: 19150886 Free PMC article.
-
Role of Notch signaling pathway in gastric cancer: a meta-analysis of the literature.World J Gastroenterol. 2014 Jul 21;20(27):9191-9. doi: 10.3748/wjg.v20.i27.9191. World J Gastroenterol. 2014. PMID: 25083094 Free PMC article. Review.
-
Ligand-dependent Notch signaling in vascular formation.Adv Exp Med Biol. 2012;727:210-22. doi: 10.1007/978-1-4614-0899-4_16. Adv Exp Med Biol. 2012. PMID: 22399350 Review.
Cited by
-
Multiple strategies to improve the therapeutic efficacy of oncolytic herpes simplex virus in the treatment of glioblastoma.Oncol Lett. 2021 Jul;22(1):510. doi: 10.3892/ol.2021.12771. Epub 2021 May 3. Oncol Lett. 2021. PMID: 33986870 Free PMC article. Review.
-
Kaposi sarcoma-associated herpesvirus: immunobiology, oncogenesis, and therapy.J Clin Invest. 2016 Sep 1;126(9):3165-75. doi: 10.1172/JCI84418. Epub 2016 Sep 1. J Clin Invest. 2016. PMID: 27584730 Free PMC article. Review.
-
Productively infected murine Kaposi's sarcoma-like tumors define new animal models for studying and targeting KSHV oncogenesis and replication.PLoS One. 2014 Jan 28;9(1):e87324. doi: 10.1371/journal.pone.0087324. eCollection 2014. PLoS One. 2014. PMID: 24489895 Free PMC article.
-
Kaposi sarcoma herpesvirus promotes endothelial-to-mesenchymal transition through Notch-dependent signaling.Cancer Res. 2012 Mar 1;72(5):1157-69. doi: 10.1158/0008-5472.CAN-11-3067. Epub 2012 Jan 11. Cancer Res. 2012. PMID: 22237624 Free PMC article.
-
Unbiased Screening of Activated Receptor Tyrosine Kinases (RTKs) in Tumor Extracts Using a Mouse Phospho-RTK Array Kit.Bio Protoc. 2019 Apr 20;9(8):e3216. doi: 10.21769/bioprotoc.3216. Bio Protoc. 2019. PMID: 32864391 Free PMC article.
References
-
- Chang Y, Cesarman E, Pessin MS, et al. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science. 1994;266(5192):1865–1869. - PubMed
-
- Sturzl M, Hohenadl C, Zietz C, et al. Expression of K13/v-FLIP gene of human herpesvirus 8 and apoptosis in Kaposi's sarcoma spindle cells. J Natl Cancer Inst. 1999;91(20):1725–1733. - PubMed
-
- Sun Q, Zachariah S, Chaudhary PM. The human herpes virus 8-encoded viral FLICE-inhibitory protein induces cellular transformation via NF-kappaB activation. J Biol Chem. 2003;278(52):52437–52445. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

