Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis
- PMID: 19805022
- PMCID: PMC2754379
- DOI: 10.1073/pnas.0907095106
Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis
Abstract
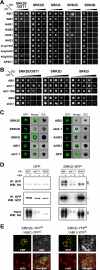
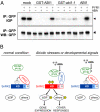
Abscisic acid (ABA) signaling is important for stress responses and developmental processes in plants. A subgroup of protein phosphatase 2C (group A PP2C) or SNF1-related protein kinase 2 (subclass III SnRK2) have been known as major negative or positive regulators of ABA signaling, respectively. Here, we demonstrate the physical and functional linkage between these two major signaling factors. Group A PP2Cs interacted physically with SnRK2s in various combinations, and efficiently inactivated ABA-activated SnRK2s via dephosphorylation of multiple Ser/Thr residues in the activation loop. This step was suppressed by the RCAR/PYR ABA receptors in response to ABA. However the abi1-1 mutated PP2C did not respond to the receptors and constitutively inactivated SnRK2. Our results demonstrate that group A PP2Cs act as 'gatekeepers' of subclass III SnRK2s, unraveling an important regulatory mechanism of ABA signaling.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Comment in
-
The PP2C-SnRK2 complex: the central regulator of an abscisic acid signaling pathway.Plant Signal Behav. 2010 Feb;5(2):160-3. doi: 10.4161/psb.5.2.10460. Epub 2010 Feb 28. Plant Signal Behav. 2010. PMID: 20023393 Free PMC article.
Similar articles
-
Phosphatase ABI1 and okadaic acid-sensitive phosphoprotein phosphatases inhibit salt stress-activated SnRK2.4 kinase.BMC Plant Biol. 2016 Jun 13;16(1):136. doi: 10.1186/s12870-016-0817-1. BMC Plant Biol. 2016. PMID: 27297076 Free PMC article.
-
PYR/PYL/RCAR family members are major in-vivo ABI1 protein phosphatase 2C-interacting proteins in Arabidopsis.Plant J. 2010 Jan;61(2):290-9. doi: 10.1111/j.1365-313X.2009.04054.x. Epub 2009 Oct 26. Plant J. 2010. PMID: 19874541 Free PMC article.
-
Protein phosphatases 2C regulate the activation of the Snf1-related kinase OST1 by abscisic acid in Arabidopsis.Plant Cell. 2009 Oct;21(10):3170-84. doi: 10.1105/tpc.109.069179. Epub 2009 Oct 23. Plant Cell. 2009. PMID: 19855047 Free PMC article.
-
Early abscisic acid signal transduction mechanisms: newly discovered components and newly emerging questions.Genes Dev. 2010 Aug 15;24(16):1695-708. doi: 10.1101/gad.1953910. Genes Dev. 2010. PMID: 20713515 Free PMC article. Review.
-
Structural insights into PYR/PYL/RCAR ABA receptors and PP2Cs.Plant Sci. 2012 Jan;182:3-11. doi: 10.1016/j.plantsci.2010.11.014. Epub 2010 Dec 7. Plant Sci. 2012. PMID: 22118610 Review.
Cited by
-
N6-methyladenosine RNA modification regulates strawberry fruit ripening in an ABA-dependent manner.Genome Biol. 2021 Jun 3;22(1):168. doi: 10.1186/s13059-021-02385-0. Genome Biol. 2021. PMID: 34078442 Free PMC article.
-
Calcium specificity signaling mechanisms in abscisic acid signal transduction in Arabidopsis guard cells.Elife. 2015 Jul 20;4:e03599. doi: 10.7554/eLife.03599. Elife. 2015. PMID: 26192964 Free PMC article.
-
Characterization of the complex regulation of AtALMT1 expression in response to phytohormones and other inducers.Plant Physiol. 2013 Jun;162(2):732-40. doi: 10.1104/pp.113.218065. Epub 2013 Apr 26. Plant Physiol. 2013. PMID: 23624855 Free PMC article.
-
Global Transcriptome Profiles of 'Meyer' Zoysiagrass in Response to Cold Stress.PLoS One. 2015 Jun 26;10(6):e0131153. doi: 10.1371/journal.pone.0131153. eCollection 2015. PLoS One. 2015. PMID: 26115186 Free PMC article.
-
A plasma membrane receptor kinase, GHR1, mediates abscisic acid- and hydrogen peroxide-regulated stomatal movement in Arabidopsis.Plant Cell. 2012 Jun;24(6):2546-61. doi: 10.1105/tpc.112.100107. Epub 2012 Jun 22. Plant Cell. 2012. PMID: 22730405 Free PMC article.
References
-
- Yamaguchi-Shinozaki K, Shinozaki K. Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu Rev Plant Biol. 2006;57:781–803. - PubMed
-
- Sirichandra C, Wasilewska A, Vlad F, Valon C, Leung J. The guard cell as a single-cell model towards understanding drought tolerance and abscisic acid action. J Exp Bot. 2009;60:1439–1463. - PubMed
-
- Finkelstein R, Reeves W, Ariizumi T, Steber C. Molecular aspects of seed dormancy. Annu Rev Plant Biol. 2008;59:387–415. - PubMed
-
- Mauch-Mani B, Mauch F. The role of abscisic acid in plant-pathogen interactions. Curr Opin Plant Biol. 2005;8:409–414. - PubMed
-
- Hirayama T, Shinozaki K. Perception and transduction of abscisic acid signals: Keys to the function of the versatile plant hormone ABA. Trends Plants Sci. 2007;12:343–351. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

