Hypoxia-inducible mir-210 regulates normoxic gene expression involved in tumor initiation
- PMID: 19782034
- PMCID: PMC2782615
- DOI: 10.1016/j.molcel.2009.09.006
Hypoxia-inducible mir-210 regulates normoxic gene expression involved in tumor initiation
Abstract
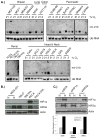
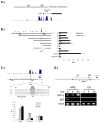
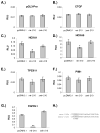
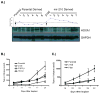
Previous studies have suggested that the HIF transcription factors can both activate and inhibit gene expression. Here we show that HIF1 regulates the expression of mir-210 in a variety of tumor types through a hypoxia-responsive element. Expression analysis in primary head and neck tumor samples indicates that mir-210 may serve as an in vivo marker for tumor hypoxia. By Argonaute protein immunoprecipitation, we identified 50 potential mir-210 targets and validated randomly selected ones. The majority of these 50 genes are not classical hypoxia-inducible genes, suggesting mir-210 represses genes expressed under normoxia that are no longer necessary to adapt and survive in a hypoxic environment. When human head and neck or pancreatic tumor cells ectopically expressing mir-210 were implanted into immunodeficient mice, mir-210 repressed initiation of tumor growth. Taken together, these data implicate an important role for mir-210 in regulating the hypoxic response of tumor cells and tumor growth.
Figures



Comment in
-
mir-210: a sensor for hypoxic stress during tumorigenesis.Mol Cell. 2009 Sep 24;35(6):737-8. doi: 10.1016/j.molcel.2009.09.008. Mol Cell. 2009. PMID: 19782023 Free PMC article.
Similar articles
-
mir-210: a sensor for hypoxic stress during tumorigenesis.Mol Cell. 2009 Sep 24;35(6):737-8. doi: 10.1016/j.molcel.2009.09.008. Mol Cell. 2009. PMID: 19782023 Free PMC article.
-
Induction, modulation and potential targets of miR-210 in pancreatic cancer cells.Hepatobiliary Pancreat Dis Int. 2012 Jun;11(3):319-24. doi: 10.1016/s1499-3872(12)60168-4. Hepatobiliary Pancreat Dis Int. 2012. PMID: 22672828
-
MiR-548an, Transcriptionally Downregulated by HIF1α/HDAC1, Suppresses Tumorigenesis of Pancreatic Cancer by Targeting Vimentin Expression.Mol Cancer Ther. 2016 Sep;15(9):2209-19. doi: 10.1158/1535-7163.MCT-15-0877. Epub 2016 Jun 28. Mol Cancer Ther. 2016. PMID: 27353169
-
The dual role of MiR-210 in the aetiology of cancer: A focus on hypoxia-inducible factor signalling.Pathol Res Pract. 2024 Jan;253:155018. doi: 10.1016/j.prp.2023.155018. Epub 2023 Dec 6. Pathol Res Pract. 2024. PMID: 38070222 Review.
-
Hypoxia-regulated microRNAs in human cancer.Acta Pharmacol Sin. 2013 Mar;34(3):336-41. doi: 10.1038/aps.2012.195. Epub 2013 Feb 4. Acta Pharmacol Sin. 2013. PMID: 23377548 Free PMC article. Review.
Cited by
-
The biological kinship of hypoxia with CSC and EMT and their relationship with deregulated expression of miRNAs and tumor aggressiveness.Biochim Biophys Acta. 2012 Dec;1826(2):272-96. doi: 10.1016/j.bbcan.2012.04.008. Epub 2012 May 10. Biochim Biophys Acta. 2012. PMID: 22579961 Free PMC article. Review.
-
HIF-1α and miR-210 differential and lineage-specific expression in systemic lupus erythematosus.Mol Immunol. 2021 May;133:128-134. doi: 10.1016/j.molimm.2021.02.019. Epub 2021 Feb 28. Mol Immunol. 2021. PMID: 33657462 Free PMC article.
-
Hypoxia-inducible miR-182 enhances HIF1α signaling via targeting PHD2 and FIH1 in prostate cancer.Sci Rep. 2015 Jul 24;5:12495. doi: 10.1038/srep12495. Sci Rep. 2015. PMID: 26205124 Free PMC article.
-
MicroRNA Expression Profiling in Clear Cell Renal Cell Carcinoma: Identification and Functional Validation of Key miRNAs.PLoS One. 2015 May 4;10(5):e0125672. doi: 10.1371/journal.pone.0125672. eCollection 2015. PLoS One. 2015. PMID: 25938468 Free PMC article.
-
CDK7 and miR-210 Co-regulate Cell-Cycle Progression of Neural Progenitors in the Developing Neocortex.Stem Cell Reports. 2016 Jul 12;7(1):69-79. doi: 10.1016/j.stemcr.2016.06.005. Stem Cell Reports. 2016. PMID: 27411104 Free PMC article.
References
-
- Bartel DP. MicroRNAs: Genomics, Biogenesis, Mechanism, and Function. Cell. 2004;116:281–297. - PubMed
-
- Beitzinger M, Peters L, Zhu JY, Kremmer E, Meister G. Identification of human microRNA targets from isolated argonaute protein complexes. RNA Biology. 2007;4:76–84. - PubMed
-
- Camps C, Buffa FM, Colella S, Moore J, Sotiriou C, Sheldon H, Harris AL, Gleadle JM, Ragoussis J. hsa-miR-210 Is Induced by Hypoxia and Is an Independent Prognostic Factor in Breast Cancer. Clin Cancer Res. 2008;14:1340–1348. - PubMed
-
- Carmeliet P, Dor Y, Herbert JM, Fukumura D, Brusselmans K, Dewerchin M, Neeman M, Bono F, Abramovitch R, Maxwell P, et al. Role of HIF- 1[alpha] in hypoxia-mediated apoptosis, cell proliferation and tumour angiogenesis. Nature. 1998;394:485–490. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

