RelB and RelE of Escherichia coli form a tight complex that represses transcription via the ribbon-helix-helix motif in RelB
- PMID: 19747491
- PMCID: PMC2812701
- DOI: 10.1016/j.jmb.2009.09.006
RelB and RelE of Escherichia coli form a tight complex that represses transcription via the ribbon-helix-helix motif in RelB
Abstract
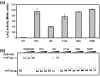
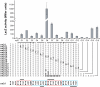
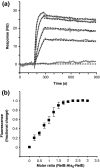
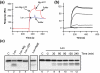
RelB, the ribbon-helix-helix (RHH) repressor encoded by the relBE toxin-antitoxin locus of Escherichia coli, interacts with RelE and thereby counteracts the mRNA cleavage activity of RelE. In addition, RelB dimers repress the strong relBE promoter and this repression by RelB is enhanced by RelE; that is, RelE functions as a transcriptional co-repressor. RelB is a Lon protease substrate, and Lon is required both for activation of relBE transcription and for activation of the mRNA cleavage activity of RelE. Here we characterize the molecular interactions important for transcriptional control of the relBE model operon. Using an in vivo screen for relB mutants, we identified multiple nucleotide changes that map to important amino acid positions within the DNA-binding domain formed by the N-terminal RHH motif of RelB. Analysis of DNA binding of a subset of these mutant RHH proteins by gel-shift assays, transcriptional fusion assays and a structure model of RelB-DNA revealed amino acid residues making crucial DNA-backbone contacts within the operator (relO) DNA. Mutational and footprinting analyses of relO showed that RelB dimers bind on the same face of the DNA helix and that the RHH motif recognizes four 6-bp repeats within the bipartite binding site. The spacing between each half-site was found to be essential for cooperative interactions between adjacently bound RelB dimers stabilized by the co-repressor RelE. Kinetic and stoichiometric measurements of the interaction between RelB and RelE confirmed that the proteins form a high-affinity complex with a 2:1 stoichiometry. Lon degraded RelB in vitro and degradation was inhibited by RelE, consistent with the proposal that RelE protects RelB from proteolysis by Lon in vivo.
Figures


Similar articles
-
Messenger RNA interferase RelE controls relBE transcription by conditional cooperativity.Mol Microbiol. 2008 Aug;69(4):841-57. doi: 10.1111/j.1365-2958.2008.06313.x. Epub 2008 Jun 4. Mol Microbiol. 2008. PMID: 18532983
-
Structural mechanism of transcriptional autorepression of the Escherichia coli RelB/RelE antitoxin/toxin module.J Mol Biol. 2008 Jun 27;380(1):107-19. doi: 10.1016/j.jmb.2008.04.039. Epub 2008 Apr 22. J Mol Biol. 2008. PMID: 18501926
-
The crystal structure of the intact E. coli RelBE toxin-antitoxin complex provides the structural basis for conditional cooperativity.Structure. 2012 Oct 10;20(10):1641-8. doi: 10.1016/j.str.2012.08.017. Epub 2012 Sep 13. Structure. 2012. PMID: 22981948 Free PMC article.
-
Structure and function of bacterial kid-kis and related toxin-antitoxin systems.Protein Pept Lett. 2007;14(2):113-24. doi: 10.2174/092986607779816096. Protein Pept Lett. 2007. PMID: 17305597 Review.
-
The Copper Efflux Regulator (CueR).Subcell Biochem. 2024;104:17-31. doi: 10.1007/978-3-031-58843-3_2. Subcell Biochem. 2024. PMID: 38963481 Review.
Cited by
-
Regulation of the Escherichia coli HipBA toxin-antitoxin system by proteolysis.PLoS One. 2012;7(6):e39185. doi: 10.1371/journal.pone.0039185. Epub 2012 Jun 15. PLoS One. 2012. PMID: 22720069 Free PMC article.
-
Hypothesis: type I toxin-antitoxin genes enter the persistence field-a feedback mechanism explaining membrane homoeostasis.Philos Trans R Soc Lond B Biol Sci. 2016 Nov 5;371(1707):20160189. doi: 10.1098/rstb.2016.0189. Philos Trans R Soc Lond B Biol Sci. 2016. PMID: 27672159 Free PMC article. Review.
-
Toxin release by conditional remodelling of ParDE1 from Mycobacterium tuberculosis leads to gyrase inhibition.Nucleic Acids Res. 2024 Feb 28;52(4):1909-1929. doi: 10.1093/nar/gkad1220. Nucleic Acids Res. 2024. PMID: 38113275 Free PMC article.
-
Three new RelE-homologous mRNA interferases of Escherichia coli differentially induced by environmental stresses.Mol Microbiol. 2010 Jan;75(2):333-48. doi: 10.1111/j.1365-2958.2009.06969.x. Epub 2009 Nov 25. Mol Microbiol. 2010. PMID: 19943910 Free PMC article.
-
Bacterial toxin RelE: a highly efficient ribonuclease with exquisite substrate specificity using atypical catalytic residues.Biochemistry. 2013 Dec 3;52(48):8633-42. doi: 10.1021/bi401325c. Epub 2013 Nov 19. Biochemistry. 2013. PMID: 24251350 Free PMC article.
References
-
- Gerdes K., Christensen S.K., Lobner-Olesen A. Prokaryotic toxin–antitoxin stress response loci. Nat. Rev. Microbiol. 2005;3:371–382. - PubMed
-
- Van Melderen L., Bernard P., Couturier M. Lon-dependent proteolysis of CcdA is the key control for activation of CcdB in plasmid-free segregant bacteria. Mol. Microbiol. 1994;11:1151–1157. - PubMed
-
- Kamada K., Hanaoka F. Conformational change in the catalytic site of the ribonuclease YoeB toxin by YefM antitoxin. Mol. Cell. 2005;19:497–509. - PubMed
-
- Pedersen K., Zavialov A.V., Pavlov M.Y., Elf J., Gerdes K., Ehrenberg M. The bacterial toxin RelE displays codon-specific cleavage of mRNAs in the ribosomal A site. Cell. 2003;112:131–140. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

