Epstein-barr virus latency in B cells leads to epigenetic repression and CpG methylation of the tumour suppressor gene Bim
- PMID: 19557159
- PMCID: PMC2695769
- DOI: 10.1371/journal.ppat.1000492
Epstein-barr virus latency in B cells leads to epigenetic repression and CpG methylation of the tumour suppressor gene Bim
Abstract
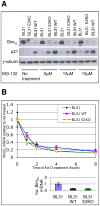
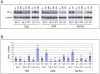
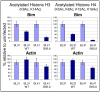
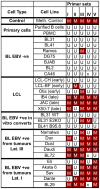
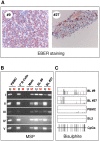
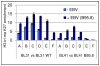
In human B cells infected with Epstein-Barr virus (EBV), latency-associated virus gene products inhibit expression of the pro-apoptotic Bcl-2-family member Bim and enhance cell survival. This involves the activities of the EBV nuclear proteins EBNA3A and EBNA3C and appears to be predominantly directed at regulating Bim mRNA synthesis, although post-transcriptional regulation of Bim has been reported. Here we show that protein and RNA stability make little or no contribution to the EBV-associated repression of Bim in latently infected B cells. However, treatment of cells with inhibitors of histone deacetylase (HDAC) and DNA methyltransferase (DNMT) enzymes indicated that epigenetic mechanisms are involved in the down-regulation of Bim. This was initially confirmed by chromatin immunoprecipitation analysis of histone acetylation levels on the Bim promoter. Consistent with this, methylation-specific PCR (MSP) and bisulphite sequencing of regions within the large CpG island located at the 5' end of Bim revealed significant methylation of CpG dinucleotides in all EBV-positive, but not EBV-negative B cells examined. Genomic DNA samples exhibiting methylation of the Bim promoter included extracts from a series of explanted EBV-positive Burkitt's lymphoma (BL) biopsies. Subsequent analyses of the histone modification H3K27-Me3 (trimethylation of histone H3 lysine 27) and CpG methylation at loci throughout the Bim promoter suggest that in EBV-positive B cells repression of Bim is initially associated with this repressive epigenetic histone mark gradually followed by DNA methylation at CpG dinucleotides. We conclude that latent EBV initiates a chain of events that leads to epigenetic repression of the tumour suppressor gene Bim in infected B cells and their progeny. This reprogramming of B cells could have important implications for our understanding of EBV persistence and the pathogenesis of EBV-associated disease, in particular BL.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




Similar articles
-
Two Epstein-Barr virus (EBV) oncoproteins cooperate to repress expression of the proapoptotic tumour-suppressor Bim: clues to the pathogenesis of Burkitt's lymphoma.Oncogene. 2008 Jan 17;27(4):421-33. doi: 10.1038/sj.onc.1210668. Epub 2007 Jul 23. Oncogene. 2008. PMID: 17653091
-
BIM promoter directly targeted by EBNA3C in polycomb-mediated repression by EBV.Nucleic Acids Res. 2012 Aug;40(15):7233-46. doi: 10.1093/nar/gks391. Epub 2012 May 14. Nucleic Acids Res. 2012. PMID: 22584624 Free PMC article.
-
The 5' regulatory sequences of active miR-146a promoters are hypomethylated and associated with euchromatic histone modification marks in B lymphoid cells.Biochem Biophys Res Commun. 2013 Apr 19;433(4):489-95. doi: 10.1016/j.bbrc.2013.03.022. Epub 2013 Mar 23. Biochem Biophys Res Commun. 2013. PMID: 23528241
-
Gene methylation in gastric cancer.Clin Chim Acta. 2013 Sep 23;424:53-65. doi: 10.1016/j.cca.2013.05.002. Epub 2013 May 10. Clin Chim Acta. 2013. PMID: 23669186 Review.
-
Epigenetic dysregulation of the host cell genome in Epstein-Barr virus-associated neoplasia.Semin Cancer Biol. 2009 Jun;19(3):158-64. doi: 10.1016/j.semcancer.2009.02.012. Epub 2009 Feb 24. Semin Cancer Biol. 2009. PMID: 19429479 Review.
Cited by
-
Epigenetic silencing of Bim transcription by Spi-1/PU.1 promotes apoptosis resistance in leukaemia.Cell Death Differ. 2013 Sep;20(9):1268-78. doi: 10.1038/cdd.2013.88. Epub 2013 Jul 12. Cell Death Differ. 2013. PMID: 23852375 Free PMC article.
-
Role of EBNA-3 Family Proteins in EBV Associated B-cell Lymphomagenesis.Front Microbiol. 2016 Apr 7;7:457. doi: 10.3389/fmicb.2016.00457. eCollection 2016. Front Microbiol. 2016. PMID: 27092119 Free PMC article. Review.
-
Expression of dual-specificity phosphatase 5 pseudogene 1 (DUSP5P1) in tumor cells.PLoS One. 2014 Feb 24;9(2):e89577. doi: 10.1371/journal.pone.0089577. eCollection 2014. PLoS One. 2014. PMID: 24651368 Free PMC article.
-
Inactivation of intergenic enhancers by EBNA3A initiates and maintains polycomb signatures across a chromatin domain encoding CXCL10 and CXCL9.PLoS Pathog. 2013 Sep;9(9):e1003638. doi: 10.1371/journal.ppat.1003638. Epub 2013 Sep 19. PLoS Pathog. 2013. PMID: 24068939 Free PMC article.
-
Epstein-Barr virus nuclear antigen 3A protein regulates CDKN2B transcription via interaction with MIZ-1.Nucleic Acids Res. 2014 Sep;42(15):9700-16. doi: 10.1093/nar/gku697. Epub 2014 Aug 4. Nucleic Acids Res. 2014. PMID: 25092922 Free PMC article.
References
-
- Thorley-Lawson DA, Gross A. Persistence of the Epstein-Barr virus and the origins of associated lymphomas. N Engl J Med. 2004;350:1328–1337. - PubMed
-
- Anderton E, Yee J, Smith P, Crook T, White RE, et al. Two Epstein-Barr virus (EBV) oncoproteins cooperate to repress expression of the proapoptotic tumour-suppressor Bim: clues to the pathogenesis of Burkitt's lymphoma. Oncogene. 2007;27:421–433. - PubMed
-
- Strasser A. The role of BH3-only proteins in the immune system. Nat Rev Immunol. 2005;5:189–200. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

