Epstein-Barr virus nuclear antigen 3C targets p53 and modulates its transcriptional and apoptotic activities
- PMID: 19394062
- PMCID: PMC4287381
- DOI: 10.1016/j.virol.2009.03.027
Epstein-Barr virus nuclear antigen 3C targets p53 and modulates its transcriptional and apoptotic activities
Abstract
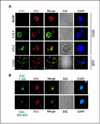
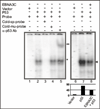
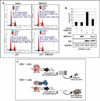
The p53 tumor suppressor gene is one of the most commonly mutated genes in human cancers and the corresponding encoded protein induces apoptosis or cell-cycle arrest at the G1/S checkpoint in response to DNA damage. To date, previous studies have shown that antigens encoded by human tumor viruses such as SV40 large T antigen, adenovirus E1A and HPV E6 interact with p53 and disrupt its functional activity. In a similar fashion, we now show that EBNA3C, one of the EBV latent antigens essential for the B-cell immortalization in vitro, interacts directly with p53. Additionally, we mapped the interaction of EBNA3C with p53 to the C-terminal DNA-binding and the tetramerization domain of p53, and the region of EBNA3C responsible for binding to p53 was mapped to the N-terminal domain of EBNA3C (residues 130-190), previously shown to interact with a number of important cell-cycle components, specifically SCF(Skp2), cyclin A, and cMyc. Furthermore, we demonstrate that EBNA3C substantially represses the transcriptional activity of p53 in luciferase based reporter assays, and rescues apoptosis induced by ectopic p53 expression in SAOS-2 (p53(-/-)) cells. Interestingly, we also show that the DNA-binding ability of p53 is diminished in the presence of EBNA3C. Thus, the interaction between the p53 and EBNA3C provides new insights into the mechanism(s) by which the EBNA3C oncoprotein can alter cellular gene expression in EBV associated human cancers.
Figures




Similar articles
-
Epstein-Barr virus nuclear antigen 3C augments Mdm2-mediated p53 ubiquitination and degradation by deubiquitinating Mdm2.J Virol. 2009 May;83(9):4652-69. doi: 10.1128/JVI.02408-08. Epub 2009 Feb 25. J Virol. 2009. PMID: 19244339 Free PMC article.
-
Epstein-Barr virus nuclear antigen 3C stabilizes Gemin3 to block p53-mediated apoptosis.PLoS Pathog. 2011 Dec;7(12):e1002418. doi: 10.1371/journal.ppat.1002418. Epub 2011 Dec 8. PLoS Pathog. 2011. Retraction in: PLoS Pathog. 2021 Apr 30;17(4):e1009556. doi: 10.1371/journal.ppat.1009556 PMID: 22174681 Free PMC article. Retracted.
-
EBNA3C attenuates the function of p53 through interaction with inhibitor of growth family proteins 4 and 5.J Virol. 2011 Mar;85(5):2079-88. doi: 10.1128/JVI.02279-10. Epub 2010 Dec 22. J Virol. 2011. PMID: 21177815 Free PMC article.
-
The Epstein Barr nuclear antigen EBNA3C regulates transcription, cell transformation and cell migration.Front Biosci. 2002 Mar 1;7:d704-16. doi: 10.2741/subraman. Front Biosci. 2002. PMID: 11861219 Review.
-
Nucleoside diphosphate kinase/Nm23 and Epstein-Barr virus.Mol Cell Biochem. 2009 Sep;329(1-2):131-9. doi: 10.1007/s11010-009-0123-4. Epub 2009 May 3. Mol Cell Biochem. 2009. PMID: 19412732 Free PMC article. Review.
Cited by
-
Epstein - Barr virus transforming protein LMP-1 alters B cells gene expression by promoting accumulation of the oncoprotein ΔNp73α.PLoS Pathog. 2013 Mar;9(3):e1003186. doi: 10.1371/journal.ppat.1003186. Epub 2013 Mar 14. PLoS Pathog. 2013. PMID: 23516355 Free PMC article.
-
Epstein-Barr virus-associated B-cell lymphomas: pathogenesis and clinical outcomes.Clin Cancer Res. 2011 May 15;17(10):3056-63. doi: 10.1158/1078-0432.CCR-10-2578. Epub 2011 Mar 3. Clin Cancer Res. 2011. PMID: 21372216 Free PMC article. Review.
-
EBNA3C-mediated regulation of aurora kinase B contributes to Epstein-Barr virus-induced B-cell proliferation through modulation of the activities of the retinoblastoma protein and apoptotic caspases.J Virol. 2013 Nov;87(22):12121-38. doi: 10.1128/JVI.02379-13. Epub 2013 Aug 28. J Virol. 2013. PMID: 23986604 Free PMC article.
-
Dissecting the contribution of EBNA3C domains important for EBV-induced B-cell growth and proliferation.Oncotarget. 2015 Oct 6;6(30):30115-29. doi: 10.18632/oncotarget.5002. Oncotarget. 2015. PMID: 26336822 Free PMC article.
-
EBV and not HPV sensitizes tobacco-associated head and neck cancer cell line FaDu to radiotherapy.Acta Otolaryngol. 2016;136(4):354-62. doi: 10.3109/00016489.2015.1114182. Epub 2015 Dec 4. Acta Otolaryngol. 2016. PMID: 26635065 Free PMC article.
References
-
- Adimoolam S, Ford JM. p53 and regulation of DNA damage recognition during nucleotide excision repair. DNA Repair. 2003;2(9):947–954. - PubMed
-
- Aiello L, Guilfoyle R, Huebner K, Weinmann R. Adenovirus 5 DNA sequences present and RNA sequences transcribed in transformed human embryo kidney cells (HEK-Ad-5 or 293) Virology. 1979;94(2):460–469. - PubMed
-
- Ambinder RF. Human lymphotropic viruses associated with lymphoid malignancy: Epstein-Barr and HTLV-1. Hematol Oncol Clin North Am. 1990;4(4):821–833. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

