Characterization of cell-surface prion protein relative to its recombinant analogue: insights from molecular dynamics simulations of diglycosylated, membrane-bound human prion protein
- PMID: 19226372
- PMCID: PMC2674387
- DOI: 10.1111/j.1471-4159.2009.05892.x
Characterization of cell-surface prion protein relative to its recombinant analogue: insights from molecular dynamics simulations of diglycosylated, membrane-bound human prion protein
Abstract
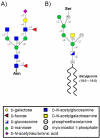
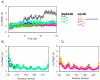
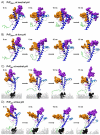
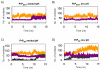
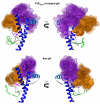
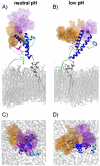
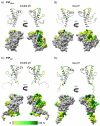
The prion protein (PrP) is responsible for several fatal neurodegenerative diseases via conversion from its normal to disease-related isoform. The recombinant form of the protein is typically studied to investigate the conversion process. This constructs lacks the co- and post-translational modifications present in vivo, there the protein has two N-linked glycans and is bound to the outer leaflet of the plasma membrane via a glycosylphosphatidylinositol (GPI) anchor. The inherent flexibility and heterogeneity of the glycans, the plasticity of the GPI anchor, and the localization of the protein in a membrane make experimental structural characterization of biological constructs of cellular prion protein (PrP(C)) challenging. Yet this characterization is central in determining not only the suitability of recombinant (rec)-PrP(C) as a model for biological forms of the protein but also the potential role of co- and post-translational modifications on the disease process. Here, we present molecular dynamics simulations of three human prion protein constructs: (i) a protein-only construct modeling the recombinant form, (ii) a diglycosylated and soluble construct, and (iii) a diglycosylated and GPI-anchored construct bound to a lipid bilayer. We found that glycosylation and membrane anchoring do not significantly alter the structure or dynamics of PrP(C), but they do appreciably modify the accessibility of the polypeptide surface PrP(C). In addition, the simulations of membrane-bound PrP(C) revealed likely recognition domains for the disease-initiating PrP(C):PrP(Sc) (infectious and/or misfolded form of the prion protein) binding event and a potential mechanism for the observed inefficiency of conversion associated with differentially glycosylated PrP species.
Figures

Similar articles
-
Simulations of membrane-bound diglycosylated human prion protein reveal potential protective mechanisms against misfolding.J Neurochem. 2017 Jul;142(1):171-182. doi: 10.1111/jnc.14044. Epub 2017 May 22. J Neurochem. 2017. PMID: 28407243 Free PMC article.
-
Cytoskeleton-dependent clustering of membrane-bound prion protein on the cell surface.J Biol Chem. 2021 Jan-Jun;296:100359. doi: 10.1016/j.jbc.2021.100359. Epub 2021 Feb 2. J Biol Chem. 2021. PMID: 33539927 Free PMC article.
-
Synthesis and structural characterization of a mimetic membrane-anchored prion protein.FEBS J. 2006 Mar;273(6):1285-99. doi: 10.1111/j.1742-4658.2006.05152.x. FEBS J. 2006. PMID: 16519692
-
Evolving views in prion glycosylation: functional and pathological implications.Biochimie. 2003 Jan-Feb;85(1-2):33-45. doi: 10.1016/s0300-9084(03)00040-3. Biochimie. 2003. PMID: 12765773 Review.
-
Conformational conversion of prion protein in prion diseases.Acta Biochim Biophys Sin (Shanghai). 2013 Jun;45(6):465-76. doi: 10.1093/abbs/gmt027. Epub 2013 Apr 11. Acta Biochim Biophys Sin (Shanghai). 2013. PMID: 23580591 Review.
Cited by
-
The consequences of pathogenic mutations to the human prion protein.Protein Eng Des Sel. 2009 Aug;22(8):461-8. doi: 10.1093/protein/gzp039. Epub 2009 Jul 14. Protein Eng Des Sel. 2009. PMID: 19602567 Free PMC article. Review.
-
The role of Cys179-Cys214 disulfide bond in the stability and folding of prion protein: insights from molecular dynamics simulations.J Mol Model. 2014 Feb;20(2):2106. doi: 10.1007/s00894-014-2106-y. Epub 2014 Feb 11. J Mol Model. 2014. PMID: 24515720
-
Molecular dynamics simulations capture the misfolding of the bovine prion protein at acidic pH.Biomolecules. 2014 Feb 10;4(1):181-201. doi: 10.3390/biom4010181. Biomolecules. 2014. PMID: 24970211 Free PMC article.
-
Protein-lipid interactions and protein anchoring modulate the modes of association of the globular domain of the Prion protein and Doppel protein to model membrane patches.Front Bioinform. 2024 Jan 5;3:1321287. doi: 10.3389/fbinf.2023.1321287. eCollection 2023. Front Bioinform. 2024. PMID: 38250434 Free PMC article.
-
Recruitment of cellular prion protein to mitochondrial raft-like microdomains contributes to apoptosis execution.Mol Biol Cell. 2011 Dec;22(24):4842-53. doi: 10.1091/mbc.E11-04-0348. Epub 2011 Oct 26. Mol Biol Cell. 2011. PMID: 22031292 Free PMC article.
References
-
- Alonso DOV, An C, Daggett V. Simulations of biomolecules: characterization of the early steps in the pH-induced conformational conversion of the hamster, bovine and human forms of the prion protein. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2002;360:1165–1178. - PubMed
-
- Baron GS, Caughey B. Effect of glycosylphosphatidylinositol anchor-dependent and-independent prion protein association with model raft membranes on conversion to the protease-resistant isoform. J. Biol. Chem. 2003;278:14883–14892. - PubMed
-
- Beck DAC, Daggett V. Methods for molecular dynamics simulations of protein folding/unfolding in solution. Methods. 2004;34:112–120. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

