Puma and to a lesser extent Noxa are suppressors of Myc-induced lymphomagenesis
- PMID: 19148184
- PMCID: PMC2743939
- DOI: 10.1038/cdd.2008.195
Puma and to a lesser extent Noxa are suppressors of Myc-induced lymphomagenesis
Abstract
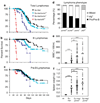
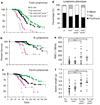
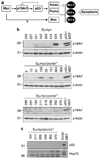
Evasion of apoptosis contributes importantly to c-Myc-induced tumorigenesis. The BH3-only Bcl-2 family members Puma and Noxa are critical pro-apoptotic transcriptional targets of p53, a major mediator of Myc-induced apoptosis and suppressor of Myc-induced tumorigenesis. Hence, we have explored the impact of their individual or combined loss on myc-driven lymphomagenesis. Notably, Puma deficiency both increased B-lineage cells and accelerated the development of B lymphoma, accompanied by leukaemia, but not of pre-B lymphoma. Noxa deficiency alone also increased B-lineage cells but did not accelerate lymphomagenesis. However, its deficiency combined with loss of one puma allele produced more rapid onset of both pre-B and B lymphomas than did loss of a single puma allele alone. Nevertheless, the acceleration evoked by loss of both genes was not as marked as that caused by p53 heterozygosity. These results show that Puma imposes a significant, and Noxa a minor barrier to c-Myc-driven lymphomagenesis. They also indicate that additional BH3-only proteins probably also drive Myc-induced apoptosis and that non-apoptotic functions of p53 may contribute substantially to its tumour suppressor role.
Conflict of interest statement
Figures





Similar articles
-
Combined loss of PUMA and p21 accelerates c-MYC-driven lymphoma development considerably less than loss of one allele of p53.Oncogene. 2016 Jul 21;35(29):3866-71. doi: 10.1038/onc.2015.457. Epub 2015 Dec 7. Oncogene. 2016. PMID: 26640149
-
Loss of a Single Mcl-1 Allele Inhibits MYC-Driven Lymphomagenesis by Sensitizing Pro-B Cells to Apoptosis.Cell Rep. 2016 Mar 15;14(10):2337-47. doi: 10.1016/j.celrep.2016.02.039. Epub 2016 Mar 3. Cell Rep. 2016. PMID: 26947081
-
Neither loss of Bik alone, nor combined loss of Bik and Noxa, accelerate murine lymphoma development or render lymphoma cells resistant to DNA damaging drugs.Cell Death Dis. 2012 May 10;3(5):e306. doi: 10.1038/cddis.2012.42. Cell Death Dis. 2012. PMID: 22573037 Free PMC article.
-
Insights about MYC and Apoptosis in B-Lymphomagenesis: An Update from Murine Models.Int J Mol Sci. 2020 Jun 15;21(12):4265. doi: 10.3390/ijms21124265. Int J Mol Sci. 2020. PMID: 32549409 Free PMC article. Review.
-
Germinal Centre B Cell Functions and Lymphomagenesis: Circuits Involving MYC and MicroRNAs.Cells. 2019 Oct 31;8(11):1365. doi: 10.3390/cells8111365. Cells. 2019. PMID: 31683676 Free PMC article. Review.
Cited by
-
p53 deficiency triggers dysregulation of diverse cellular processes in physiological oxygen.J Cell Biol. 2020 Nov 2;219(11):e201908212. doi: 10.1083/jcb.201908212. J Cell Biol. 2020. PMID: 32886745 Free PMC article.
-
Unravelling mechanisms of p53-mediated tumour suppression.Nat Rev Cancer. 2014 May;14(5):359-70. doi: 10.1038/nrc3711. Epub 2014 Apr 17. Nat Rev Cancer. 2014. PMID: 24739573 Free PMC article.
-
Regulation of the Mdm2-p53 signaling axis in the DNA damage response and tumorigenesis.Transl Cancer Res. 2016 Dec;5(6):707-724. doi: 10.21037/tcr.2016.11.75. Transl Cancer Res. 2016. PMID: 28690977 Free PMC article.
-
Dynein light chain regulates adaptive and innate B cell development by distinctive genetic mechanisms.PLoS Genet. 2017 Sep 18;13(9):e1007010. doi: 10.1371/journal.pgen.1007010. eCollection 2017 Sep. PLoS Genet. 2017. PMID: 28922373 Free PMC article.
-
The tumor-modulatory effects of Caspase-2 and Pidd1 do not require the scaffold protein Raidd.Cell Death Differ. 2015 Nov;22(11):1803-11. doi: 10.1038/cdd.2015.31. Epub 2015 Apr 10. Cell Death Differ. 2015. PMID: 25857265 Free PMC article.
References
-
- Vousden KH, Lu X. Live or let die: the cell’s response to p53. Nat Rev Cancer. 2002;2:594–604. - PubMed
-
- Kastan MB. Wild-type p53: tumors canșt stand it. Cell. 2007;128:837–840. - PubMed
-
- Wang Y, Szekely L, Okan I, Klein G, Wiman KG. Wild-type p53-triggered apoptosis is inhibited by bcl-2 in a v-myc-induced T-cell lymphoma line. Oncogene. 1993;8:3427–3431. - PubMed
-
- Strasser A, Harris AW, Cory S. Bcl-2 transgene inhibits T cell death and perturbs thymic self-censorship. Cell. 1991;67:889–899. - PubMed
-
- Strasser A, Harris AW, Jacks T, Cory S. DNA damage can induce apoptosis in proliferating lymphoid cells via p53-independent mechanisms inhibitable by Bcl-2. Cell. 1994;79:329–339. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

