The DNA-encoded nucleosome organization of a eukaryotic genome
- PMID: 19092803
- PMCID: PMC2658732
- DOI: 10.1038/nature07667
The DNA-encoded nucleosome organization of a eukaryotic genome
Abstract
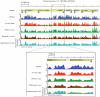
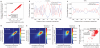
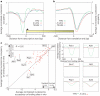
Nucleosome organization is critical for gene regulation. In living cells this organization is determined by multiple factors, including the action of chromatin remodellers, competition with site-specific DNA-binding proteins, and the DNA sequence preferences of the nucleosomes themselves. However, it has been difficult to estimate the relative importance of each of these mechanisms in vivo, because in vivo nucleosome maps reflect the combined action of all influencing factors. Here we determine the importance of nucleosome DNA sequence preferences experimentally by measuring the genome-wide occupancy of nucleosomes assembled on purified yeast genomic DNA. The resulting map, in which nucleosome occupancy is governed only by the intrinsic sequence preferences of nucleosomes, is similar to in vivo nucleosome maps generated in three different growth conditions. In vitro, nucleosome depletion is evident at many transcription factor binding sites and around gene start and end sites, indicating that nucleosome depletion at these sites in vivo is partly encoded in the genome. We confirm these results with a micrococcal nuclease-independent experiment that measures the relative affinity of nucleosomes for approximately 40,000 double-stranded 150-base-pair oligonucleotides. Using our in vitro data, we devise a computational model of nucleosome sequence preferences that is significantly correlated with in vivo nucleosome occupancy in Caenorhabditis elegans. Our results indicate that the intrinsic DNA sequence preferences of nucleosomes have a central role in determining the organization of nucleosomes in vivo.
Figures

Similar articles
-
A genomic code for nucleosome positioning.Nature. 2006 Aug 17;442(7104):772-8. doi: 10.1038/nature04979. Epub 2006 Jul 19. Nature. 2006. PMID: 16862119 Free PMC article.
-
Diversity of eukaryotic DNA replication origins revealed by genome-wide analysis of chromatin structure.PLoS Genet. 2010 Sep 2;6(9):e1001092. doi: 10.1371/journal.pgen.1001092. PLoS Genet. 2010. PMID: 20824081 Free PMC article.
-
Prediction of nucleosome positioning in genomes: limits and perspectives of physical and bioinformatic approaches.J Biomol Struct Dyn. 2010 Jun;27(6):747-64. doi: 10.1080/07391102.2010.10508583. J Biomol Struct Dyn. 2010. PMID: 20232931
-
Nucleosome positioning in yeasts: methods, maps, and mechanisms.Chromosoma. 2015 Jun;124(2):131-51. doi: 10.1007/s00412-014-0501-x. Epub 2014 Dec 23. Chromosoma. 2015. PMID: 25529773 Review.
-
Active nucleosome positioning beyond intrinsic biophysics is revealed by in vitro reconstitution.Biochem Soc Trans. 2012 Apr;40(2):377-82. doi: 10.1042/BST20110730. Biochem Soc Trans. 2012. PMID: 22435815 Review.
Cited by
-
Nucleosome spacing generated by ISWI and CHD1 remodelers is constant regardless of nucleosome density.Mol Cell Biol. 2015 May;35(9):1588-605. doi: 10.1128/MCB.01070-14. Epub 2015 Mar 2. Mol Cell Biol. 2015. PMID: 25733687 Free PMC article.
-
DNA sequence preferences of transcriptional activators correlate more strongly than repressors with nucleosomes.Mol Cell. 2012 Jul 27;47(2):183-92. doi: 10.1016/j.molcel.2012.06.028. Mol Cell. 2012. PMID: 22841002 Free PMC article.
-
The Upstream Sequence Transcription Complex dictates nucleosome positioning and promoter accessibility at piRNA genes in the C. elegans germ line.PLoS Genet. 2024 Jul 10;20(7):e1011345. doi: 10.1371/journal.pgen.1011345. eCollection 2024 Jul. PLoS Genet. 2024. PMID: 38985845 Free PMC article.
-
Drosophila functional elements are embedded in structurally constrained sequences.PLoS Genet. 2013 May;9(5):e1003512. doi: 10.1371/journal.pgen.1003512. Epub 2013 May 30. PLoS Genet. 2013. PMID: 23750124 Free PMC article.
-
Multiplexing mechanical and translational cues on genes.Biophys J. 2022 Nov 15;121(22):4311-4324. doi: 10.1016/j.bpj.2022.10.011. Epub 2022 Oct 13. Biophys J. 2022. PMID: 36230003 Free PMC article.
References
-
- Feng HP, Scherl DS, Widom J. Lifetime of the histone octamer studied by continuous-flow quasielastic light scattering: test of a model for nucleosome transcription. Biochemistry. 1993;32:7824–7831. - PubMed
-
- Porreca GJ, et al. Multiplex amplification of large sets of human exons. Nature Methods. 2007;4:931–936. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

