Toll-like receptor 4 mediates innate immunity to Kaposi sarcoma herpesvirus
- PMID: 18996347
- PMCID: PMC2698447
- DOI: 10.1016/j.chom.2008.09.012
Toll-like receptor 4 mediates innate immunity to Kaposi sarcoma herpesvirus
Abstract
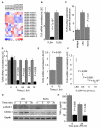
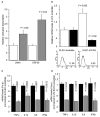
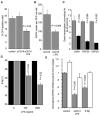
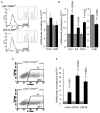
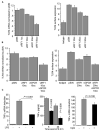
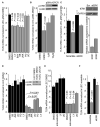
The involvement of Toll-like receptor 4 (TLR4) in immunity against human herpesviruses has not been previously demonstrated. We show that infection of endothelial cells with Kaposi sarcoma herpesvirus (KSHV), a human oncogenic virus, leads to rapid suppression of TLR4 expression. This is a mechanism of immune escape as TLR4 mediates innate immunity against KSHV. In vitro, cells lacking TLR4 are more susceptible to KSHV infection, whereas activation of TLR4 protects cells from infection. In vivo, HIV-1-infected individuals carrying a mutant TLR4 allele appear more likely to have multicentric Castleman's disease, a lymphoproliferation associated with enhanced KSHV replication. ERK activation by KSHV structural proteins and the KSHV-encoded vGPCR plays a key role in the TLR4 downregulation, whereas the KSHV vIRF1 also contributes to this effect. Our findings reveal a role for TLR4 in innate immunity against herpesviruses and suggest the potential use of TLR4 agonists for the treatment of KSHV-related neoplasms.
Figures

Similar articles
-
Pseudomonas aeruginosa Stimulates Inflammation and Enhances Kaposi's Sarcoma Herpesvirus-Induced Cell Proliferation and Cellular Transformation through both Lipopolysaccharide and Flagellin.mBio. 2020 Nov 10;11(6):e02843-20. doi: 10.1128/mBio.02843-20. mBio. 2020. PMID: 33173008 Free PMC article.
-
Kaposi sarcoma-associated herpesvirus complement control protein (KCP) and glycoprotein K8.1 are not required for viral infection in vitro or in vivo.J Virol. 2024 Jun 13;98(6):e0057624. doi: 10.1128/jvi.00576-24. Epub 2024 May 20. J Virol. 2024. PMID: 38767375 Free PMC article.
-
The gammaherpesviruses Kaposi's sarcoma-associated herpesvirus and murine gammaherpesvirus 68 modulate the Toll-like receptor-induced proinflammatory cytokine response.J Virol. 2014 Aug;88(16):9245-59. doi: 10.1128/JVI.00841-14. Epub 2014 Jun 4. J Virol. 2014. PMID: 24899179 Free PMC article.
-
Kaposi sarcoma-associated herpesvirus: immunobiology, oncogenesis, and therapy.J Clin Invest. 2016 Sep 1;126(9):3165-75. doi: 10.1172/JCI84418. Epub 2016 Sep 1. J Clin Invest. 2016. PMID: 27584730 Free PMC article. Review.
-
Interplay between Kaposi's sarcoma-associated herpesvirus and the innate immune system.Cytokine Growth Factor Rev. 2014 Oct;25(5):597-609. doi: 10.1016/j.cytogfr.2014.06.001. Epub 2014 Jun 21. Cytokine Growth Factor Rev. 2014. PMID: 25037686 Free PMC article. Review.
Cited by
-
Molecular Biology of KSHV in Relation to HIV/AIDS-Associated Oncogenesis.Cancer Treat Res. 2019;177:23-62. doi: 10.1007/978-3-030-03502-0_2. Cancer Treat Res. 2019. PMID: 30523620 Free PMC article.
-
KSHV: pathways to tumorigenesis and persistent infection.Adv Virus Res. 2014;88:111-59. doi: 10.1016/B978-0-12-800098-4.00002-7. Adv Virus Res. 2014. PMID: 24373311 Free PMC article. Review.
-
Kaposi sarcoma herpesvirus pathogenesis.Philos Trans R Soc Lond B Biol Sci. 2017 Oct 19;372(1732):20160275. doi: 10.1098/rstb.2016.0275. Philos Trans R Soc Lond B Biol Sci. 2017. PMID: 28893942 Free PMC article. Review.
-
The Heme Metabolite Carbon Monoxide Facilitates KSHV Infection by Inhibiting TLR4 Signaling in Endothelial Cells.Front Microbiol. 2017 Apr 3;8:568. doi: 10.3389/fmicb.2017.00568. eCollection 2017. Front Microbiol. 2017. PMID: 28421060 Free PMC article.
-
The Chinese Herbal Prescription JieZe-1 Inhibits Membrane Fusion and the Toll-like Receptor Signaling Pathway in a Genital Herpes Mouse Model.Front Pharmacol. 2021 Sep 24;12:707695. doi: 10.3389/fphar.2021.707695. eCollection 2021. Front Pharmacol. 2021. PMID: 34630083 Free PMC article.
References
-
- Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. - PubMed
-
- Apetoh L, Ghiringhelli F, Tesniere A, Obeid M, Ortiz C, Criollo A, Mignot G, Maiuri MC, Ullrich E, Saulnier P, et al. Toll-like receptor 4-dependent contribution of the immune system to anticancer chemotherapy and radiotherapy. Nat. Med. 2007;13:1050–1059. - PubMed
-
- Boivin G, Cote S, Cloutier N, Abed Y, Maguigad M, Routy JP. Quantification of human herpesvirus 8 by real-time PCR in blood fractions of AIDS patients with Kaposi's sarcoma and multicentric Castleman's disease. J. Med. Virol. 2002;68:399–403. - PubMed
-
- Boshoff C, Weiss R. AIDS-related malignancies. Nat. Rev. Cancer. 2002;2:373–382. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

