DNA-free RNA isolation protocols for Arabidopsis thaliana, including seeds and siliques
- PMID: 18937828
- PMCID: PMC2613888
- DOI: 10.1186/1756-0500-1-93
DNA-free RNA isolation protocols for Arabidopsis thaliana, including seeds and siliques
Abstract
Background: High throughput applications of the reverse transcriptase quantitative PCR (RT-qPCR) for quantification of gene expression demand straightforward procedures to isolate and analyze a considerable number of DNA-free RNA samples. Published protocols are labour intensive, use toxic organic chemicals and need a DNase digestion once pure RNAs have been isolated. In addition, for some tissues, the amount of starting material may be limiting. The convenience of commercial kits is often prohibitive when handling large number of samples.
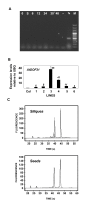
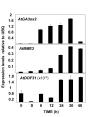
Findings: We have established protocols to isolate DNA-free RNA from Arabidopsis thaliana tissues ready for RT-qPCR applications. Simple non-toxic buffers were used for RNA isolation from Arabidopsis tissues with the exception of seeds and siliques, which required the use of organic extractions. The protocols were designed to minimize the number of steps, labour time and the amount of starting tissue to as little as 10-20 mg without affecting RNA quality. In both protocols genomic DNA (gDNA) can be efficiently removed from RNA samples before the final alcohol precipitation step, saving extra purification steps before cDNA synthesis. The expression kinetics of previously characterized genes confirmed the robustness of the procedures.
Conclusion: Here, we present two protocols to isolate DNA-free RNA from Arabidopsis tissues ready for RT-qPCR applications that significantly improve existing ones by reducing labour time and the use of organic extractions. Accessibility to these protocols is ensured by its simplicity and the low cost of the materials used.
Figures

Similar articles
-
Development of a versatile TaqMan™ real-time quantitative PCR (RT-qPCR) compliant anchor sequence to quantify bacterial gene transcripts from RNA samples containing carryover genomic DNA.BMC Biotechnol. 2013 Jan 31;13:7. doi: 10.1186/1472-6750-13-7. BMC Biotechnol. 2013. PMID: 23369378 Free PMC article.
-
A rapid TRIzol-based two-step method for DNA-free RNA extraction from Arabidopsis siliques and dry seeds.Biotechnol J. 2010 Feb;5(2):183-6. doi: 10.1002/biot.200900211. Biotechnol J. 2010. PMID: 20108272
-
Simple, Inexpensive RNA Isolation and One-Step RT-qPCR Methods for SARS-CoV-2 Detection and General Use.Curr Protoc. 2021 Apr;1(4):e130. doi: 10.1002/cpz1.130. Curr Protoc. 2021. PMID: 33905620 Free PMC article.
-
Identification of imprinted genes subject to parent-of-origin specific expression in Arabidopsis thaliana seeds.BMC Plant Biol. 2011 Aug 12;11:113. doi: 10.1186/1471-2229-11-113. BMC Plant Biol. 2011. PMID: 21838868 Free PMC article.
-
Isolation of high-quality RNA from polyphenol-, polysaccharide- and lipid-rich seeds.Phytochem Anal. 2006 May-Jun;17(3):144-8. doi: 10.1002/pca.903. Phytochem Anal. 2006. PMID: 16749420
Cited by
-
Optimization of quantitative reverse transcription PCR method for analysis of weakly expressed genes in crops based on rapeseed.Front Plant Sci. 2022 Aug 9;13:954976. doi: 10.3389/fpls.2022.954976. eCollection 2022. Front Plant Sci. 2022. PMID: 36017265 Free PMC article.
-
Mannans and endo-β-mannanases (MAN) in Brachypodium distachyon: expression profiling and possible role of the BdMAN genes during coleorhiza-limited seed germination.J Exp Bot. 2015 Jul;66(13):3753-64. doi: 10.1093/jxb/erv168. Epub 2015 Apr 28. J Exp Bot. 2015. PMID: 25922488 Free PMC article.
-
SUCROSE TRANSPORTER 5 supplies Arabidopsis embryos with biotin and affects triacylglycerol accumulation.Plant J. 2013 Feb;73(3):392-404. doi: 10.1111/tpj.12037. Epub 2012 Dec 31. Plant J. 2013. PMID: 23031218 Free PMC article.
-
Arabidopsis AL PHD-PRC1 complexes promote seed germination through H3K4me3-to-H3K27me3 chromatin state switch in repression of seed developmental genes.PLoS Genet. 2014 Jan;10(1):e1004091. doi: 10.1371/journal.pgen.1004091. Epub 2014 Jan 23. PLoS Genet. 2014. PMID: 24465219 Free PMC article.
-
The glycosyltransferase UGT76E1 significantly contributes to 12-O-glucopyranosyl-jasmonic acid formation in wounded Arabidopsis thaliana leaves.J Biol Chem. 2019 Jun 21;294(25):9858-9872. doi: 10.1074/jbc.RA119.007600. Epub 2019 May 9. J Biol Chem. 2019. PMID: 31072871 Free PMC article.
References
-
- Carpenter CD, Simon AE. Preparation of RNA. In: Martínez-Zapater JM, Salinas J, editor. Methods in Molecular Biology, Arabidopsis Protocols. Vol. 82. Totowa: Humana Press; 1998. pp. 85–89. - PubMed
-
- Weigel D, Glazebrook J. Arabidopsis: A laboratory manual. New York: Cold Spring Harbor, Cold Spring Harbor Laboratory Press; 2002.
-
- Suzuki Y, Kawazu T, Koyama H. RNA isolation from siliques, dry seeds, and other tissues of Arabidopsis thaliana. BioTechniques. 2004;37:542–544. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

