Identification of monomorphic and divergent haplotypes in the 2006-2007 norovirus GII/4 epidemic population by genomewide tracing of evolutionary history
- PMID: 18768979
- PMCID: PMC2573252
- DOI: 10.1128/JVI.00897-08
Identification of monomorphic and divergent haplotypes in the 2006-2007 norovirus GII/4 epidemic population by genomewide tracing of evolutionary history
Abstract
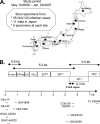
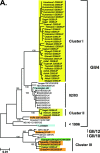
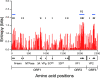
Our norovirus (NoV) surveillance group reported a >4-fold increase in NoV infection in Japan during the winter of 2006-2007 compared to the previous winter. Because the increase was not linked to changes in the surveillance system, we suspected the emergence of new NoV GII/4 epidemic variants. To obtain information on viral changes, we conducted full-length genomic analysis. Stool specimens from 55 acute gastroenteritis patients of various ages were collected at 11 sites in Japan between May 2006 and January 2007. Direct sequencing of long PCR products revealed 37 GII/4 genome sequences. Phylogenetic study of viral genome and partial sequences showed that the two new GII/4 variants in Europe, termed 2006a and 2006b, initially coexisted as minorities in early 2006 in Japan and that 2006b alone had dominated over the resident GII/4 variants during 2006. A combination of phylogenetic and entropy analyses revealed for the first time the unique amino acid substitutions in all eight proteins of the new epidemic strains. These data and computer-assisted structural study of the NoV capsid protein are compatible with a model of antigenic drift with tuning of the structure and functions of multiple proteins for the global outgrowth of new GII/4 variants. The availability of comprehensive information on genome sequences and unique protein changes of the recent global epidemic variants will allow studies of diagnostic assays, molecular epidemiology, molecular biology, and adaptive changes of NoV in nature.
Figures








Similar articles
-
Molecular epidemiology of norovirus in Singapore, 2004-2011.J Med Virol. 2013 Oct;85(10):1842-51. doi: 10.1002/jmv.23669. Epub 2013 Jul 19. J Med Virol. 2013. PMID: 23868077
-
Divergent evolution of norovirus GII/4 by genome recombination from May 2006 to February 2009 in Japan.J Virol. 2010 Aug;84(16):8085-97. doi: 10.1128/JVI.02125-09. Epub 2010 Jun 9. J Virol. 2010. PMID: 20534859 Free PMC article.
-
Temporal evolutionary analysis of re-emerging recombinant GII.P16_GII.2 norovirus with acute gastroenteritis in patients from Hubei Province of China, 2017.Virus Res. 2018 Apr 2;249:99-109. doi: 10.1016/j.virusres.2018.03.016. Epub 2018 Mar 28. Virus Res. 2018. PMID: 29604360
-
Emergence of a new norovirus GII.6 variant in Japan, 2008-2009.J Med Virol. 2012 Jul;84(7):1089-96. doi: 10.1002/jmv.23309. J Med Virol. 2012. PMID: 22585727
-
Clinical relevance and genotypes of circulating noroviruses in northern Taiwan, 2006-2011.J Med Virol. 2014 Feb;86(2):335-46. doi: 10.1002/jmv.23728. Epub 2013 Sep 5. J Med Virol. 2014. PMID: 24009100
Cited by
-
Molecular Evolutionary Analyses of the RNA-Dependent RNA Polymerase Region in Norovirus Genogroup II.Front Microbiol. 2018 Dec 18;9:3070. doi: 10.3389/fmicb.2018.03070. eCollection 2018. Front Microbiol. 2018. PMID: 30619155 Free PMC article.
-
Structure-Guided Creation of an Anti-HA Stalk Antibody F11 Derivative That Neutralizes Both F11-Sensitive and -Resistant Influenza A(H1N1)pdm09 Viruses.Viruses. 2021 Aug 31;13(9):1733. doi: 10.3390/v13091733. Viruses. 2021. PMID: 34578314 Free PMC article.
-
Evolutionary Constraints on the Norovirus Pandemic Variant GII.4_2006b over the Five-Year Persistence in Japan.Front Microbiol. 2017 Mar 13;8:410. doi: 10.3389/fmicb.2017.00410. eCollection 2017. Front Microbiol. 2017. PMID: 28348551 Free PMC article.
-
Molecular evolution of the VP1, VP2, and VP3 genes in human rhinovirus species C.Sci Rep. 2015 Feb 2;5:8185. doi: 10.1038/srep08185. Sci Rep. 2015. PMID: 25640899 Free PMC article.
-
Molecular Evolution of the Capsid Gene in Norovirus Genogroup I.Sci Rep. 2015 Sep 4;5:13806. doi: 10.1038/srep13806. Sci Rep. 2015. PMID: 26338545 Free PMC article.
References
-
- Ando, T., J. S. Noel, and R. L. Fankhauser. 2000. Genetic classification of “Norwalk-like viruses.” J. Infect. Dis. 181(Suppl. 2):S336-S348. - PubMed
-
- Baker, D., and A. Sali. 2001. Protein structure prediction and structural genomics. Science 29493-96. - PubMed
-
- Cauchi, M. R., J. C. Doultree, J. A. Marshall, and P. J. Wright. 1996. Molecular characterization of Camberwell virus and sequence variation in ORF3 of small round-structured (Norwalk-like) viruses. J. Med. Virol. 4970-76. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

