Viral genome methylation as an epigenetic defense against geminiviruses
- PMID: 18596098
- PMCID: PMC2546898
- DOI: 10.1128/JVI.00719-08
Viral genome methylation as an epigenetic defense against geminiviruses
Abstract
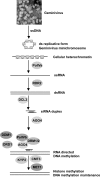
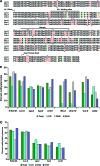
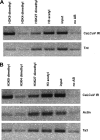
Geminiviruses encapsidate single-stranded DNA genomes that replicate in plant cell nuclei through double-stranded DNA intermediates that associate with cellular histone proteins to form minichromosomes. Like most plant viruses, geminiviruses are targeted by RNA silencing and encode suppressor proteins such as AL2 and L2 to counter this defense. These related proteins can suppress silencing by multiple mechanisms, one of which involves interacting with and inhibiting adenosine kinase (ADK), a cellular enzyme associated with the methyl cycle that generates S-adenosyl-methionine, an essential methyltransferase cofactor. Thus, we hypothesized that the viral genome is targeted by small-RNA-directed methylation. Here, we show that Arabidopsis plants with mutations in genes encoding cytosine or histone H3 lysine 9 (H3K9) methyltransferases, RNA-directed methylation pathway components, or ADK are hypersensitive to geminivirus infection. We also demonstrate that viral DNA and associated histone H3 are methylated in infected plants and that cytosine methylation levels are significantly reduced in viral DNA isolated from methylation-deficient mutants. Finally, we demonstrate that Beet curly top virus L2- mutant DNA present in tissues that have recovered from infection is hypermethylated and that host recovery requires AGO4, a component of the RNA-directed methylation pathway. We propose that plants use chromatin methylation as a defense against DNA viruses, which geminiviruses counter by inhibiting global methylation. In addition, our results establish that geminiviruses can be useful models for genome methylation in plants and suggest that there are redundant pathways leading to cytosine methylation.
Figures

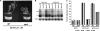
Similar articles
-
Geminivirus AL2 and L2 proteins suppress transcriptional gene silencing and cause genome-wide reductions in cytosine methylation.J Virol. 2009 May;83(10):5005-13. doi: 10.1128/JVI.01771-08. Epub 2009 Mar 11. J Virol. 2009. PMID: 19279102 Free PMC article.
-
Arabidopsis double-stranded RNA binding protein DRB3 participates in methylation-mediated defense against geminiviruses.J Virol. 2014 Mar;88(5):2611-22. doi: 10.1128/JVI.02305-13. Epub 2013 Dec 18. J Virol. 2014. PMID: 24352449 Free PMC article.
-
Arabidopsis RNA Polymerase V Mediates Enhanced Compaction and Silencing of Geminivirus and Transposon Chromatin during Host Recovery from Infection.J Virol. 2018 Mar 14;92(7):e01320-17. doi: 10.1128/JVI.01320-17. Print 2018 Apr 1. J Virol. 2018. PMID: 29321305 Free PMC article.
-
Epigenetic regulation of geminivirus pathogenesis: a case of relentless recalibration of defence responses in plants.J Exp Bot. 2020 Dec 31;71(22):6890-6906. doi: 10.1093/jxb/eraa406. J Exp Bot. 2020. PMID: 32869846 Review.
-
Protein modification: A critical modulator in the interaction between geminiviruses and host plants.Plant Cell Environ. 2021 Jun;44(6):1707-1715. doi: 10.1111/pce.14008. Epub 2021 Feb 15. Plant Cell Environ. 2021. PMID: 33506956 Review.
Cited by
-
Differential response of cassava genotypes to infection by cassava mosaic geminiviruses.Virus Res. 2017 Jan 2;227:69-81. doi: 10.1016/j.virusres.2016.09.022. Epub 2016 Sep 29. Virus Res. 2017. PMID: 27693919 Free PMC article.
-
Extreme Resistance to Viruses in Potato and Soybean.Front Plant Sci. 2021 Apr 6;12:658981. doi: 10.3389/fpls.2021.658981. eCollection 2021. Front Plant Sci. 2021. PMID: 33889169 Free PMC article. Review.
-
Tomato yellow leaf curl viruses: ménage à trois between the virus complex, the plant and the whitefly vector.Mol Plant Pathol. 2010 Jul;11(4):441-50. doi: 10.1111/j.1364-3703.2010.00618.x. Mol Plant Pathol. 2010. PMID: 20618703 Free PMC article.
-
Composted Municipal Green Waste Infused with Biocontrol Agents to Control Plant Parasitic Nematodes-A Review.Microorganisms. 2021 Oct 11;9(10):2130. doi: 10.3390/microorganisms9102130. Microorganisms. 2021. PMID: 34683451 Free PMC article. Review.
-
Involvement of host regulatory pathways during geminivirus infection: a novel platform for generating durable resistance.Funct Integr Genomics. 2014 Mar;14(1):47-58. doi: 10.1007/s10142-013-0346-z. Funct Integr Genomics. 2014. PMID: 24233104 Review.
References
-
- Baulcombe, D. C. 2004. RNA silencing in plants. Nature 431356-363. - PubMed
-
- Bender, J. 2004. DNA methylation and epigenetics. Annu. Rev. Plant Biol. 5541-68. - PubMed
-
- Bian, X.-Y., M. S. Rasheed, M. J. Seemanpillai, and M. A. Rezaian. 2006. Analysis of silencing escape of Tomato leaf curl virus: an evaluation of the role of DNA methylation. Mol. Plant-Microbe Interact. 19614-624. - PubMed
-
- Bisaro, D. M. 2006. Silencing suppression by geminivirus proteins. Virology 344158-168. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

