The evolutionary dynamics of human influenza B virus
- PMID: 18504518
- PMCID: PMC3326418
- DOI: 10.1007/s00239-008-9119-z
The evolutionary dynamics of human influenza B virus
Abstract
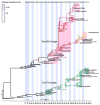
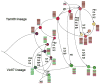
Despite their close phylogenetic relationship, type A and B influenza viruses exhibit major epidemiological differences in humans, with the latter both less common and less often associated with severe disease. However, it is unclear what processes determine the evolutionary dynamics of influenza B virus, and how influenza viruses A and B interact at the evolutionary scale. To address these questions we inferred the phylogenetic history of human influenza B virus using complete genome sequences for which the date (day) of isolation was available. By comparing the phylogenetic patterns of all eight viral segments we determined the occurrence of segment reassortment over a 30-year sampling period. An analysis of rates of nucleotide substitution and selection pressures revealed sporadic occurrences of adaptive evolution, most notably in the viral hemagglutinin and compatible with the action of antigenic drift, yet lower rates of overall and nonsynonymous nucleotide substitution compared to influenza A virus. Overall, these results led us to propose a model in which evolutionary changes within and between the antigenically distinct 'Yam88' and 'Vic87' lineages of influenza B virus are the result of changes in herd immunity, with reassortment continuously generating novel genetic variation. Additionally, we suggest that the interaction with influenza A virus may be central in shaping the evolutionary dynamics of influenza B virus, facilitating the shift of dominance between the Vic87 and the Yam88 lineages.
Figures
Similar articles
-
Genome-wide evolutionary dynamics of influenza B viruses on a global scale.PLoS Pathog. 2017 Dec 28;13(12):e1006749. doi: 10.1371/journal.ppat.1006749. eCollection 2017 Dec. PLoS Pathog. 2017. PMID: 29284042 Free PMC article.
-
The contrasting phylodynamics of human influenza B viruses.Elife. 2015 Jan 16;4:e05055. doi: 10.7554/eLife.05055. Elife. 2015. PMID: 25594904 Free PMC article.
-
Genetic diversity of influenza B virus: the frequent reassortment and cocirculation of the genetically distinct reassortant viruses in a community.J Med Virol. 2004 Sep;74(1):132-40. doi: 10.1002/jmv.20156. J Med Virol. 2004. PMID: 15258979
-
Recent changes among human influenza viruses.Virus Res. 2004 Jul;103(1-2):47-52. doi: 10.1016/j.virusres.2004.02.011. Virus Res. 2004. PMID: 15163487 Review.
-
[Molecular characterization of human influenza viruses--a look back on the last 10 years].Berl Munch Tierarztl Wochenschr. 2006 Mar-Apr;119(3-4):167-78. Berl Munch Tierarztl Wochenschr. 2006. PMID: 16573207 Review. German.
Cited by
-
Increasing serum antibodies against type B influenza virus in 2017-2018 winter in Beijing, China.AMB Express. 2022 Oct 1;12(1):127. doi: 10.1186/s13568-022-01469-9. AMB Express. 2022. PMID: 36182978 Free PMC article.
-
ADCC: An underappreciated correlate of cross-protection against influenza?Front Immunol. 2023 Feb 22;14:1130725. doi: 10.3389/fimmu.2023.1130725. eCollection 2023. Front Immunol. 2023. PMID: 36911705 Free PMC article. Review.
-
Non-Uniform and Non-Random Binding of Nucleoprotein to Influenza A and B Viral RNA.Viruses. 2018 Sep 25;10(10):522. doi: 10.3390/v10100522. Viruses. 2018. PMID: 30257455 Free PMC article.
-
Influenza B associated paediatric acute respiratory infection hospitalization in central vietnam.Influenza Other Respir Viruses. 2019 May;13(3):248-261. doi: 10.1111/irv.12626. Epub 2019 Feb 28. Influenza Other Respir Viruses. 2019. PMID: 30575288 Free PMC article.
-
Limited compatibility of polymerase subunit interactions in influenza A and B viruses.J Biol Chem. 2010 May 28;285(22):16704-12. doi: 10.1074/jbc.M110.102533. Epub 2010 Apr 2. J Biol Chem. 2010. PMID: 20363752 Free PMC article.
References
-
- Baigent SJ, McCauley JW. Influenza type A in humans, mammals and birds: determinants of virus virulence, host-range and interspecies transmission. BioEssays. 2003;25:657–671. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

