Kaposi's sarcoma-associated herpesvirus-encoded LANA can interact with the nuclear mitotic apparatus protein to regulate genome maintenance and segregation
- PMID: 18417561
- PMCID: PMC2447046
- DOI: 10.1128/JVI.00342-08
Kaposi's sarcoma-associated herpesvirus-encoded LANA can interact with the nuclear mitotic apparatus protein to regulate genome maintenance and segregation
Abstract
Kaposi's sarcoma-associated herpesvirus (KSHV) genomes are tethered to the host chromosomes and partitioned faithfully into daughter cells with the host chromosomes. The latency-associated nuclear antigen (LANA) is important for segregation of the newly synthesized viral genomes to the daughter nuclei. Here, we report that the nuclear mitotic apparatus protein (NuMA) and LANA can associate in KSHV-infected cells. In synchronized cells, NuMA and LANA are colocalized in interphase cells and separate during mitosis at the beginning of prophase, reassociating again at the end of telophase and cytokinesis. Silencing of NuMA expression by small interfering RNA and expression of LGN and a dominant-negative of dynactin (P150-CC1), which disrupts the association of NuMA with microtubules, resulted in the loss of KSHV terminal-repeat plasmids containing the major latent origin. Thus, NuMA is required for persistence of the KSHV episomes in daughter cells. This interaction between NuMA and LANA is critical for segregation and maintenance of the KSHV episomes through a temporally controlled mechanism of binding and release during specific phases of mitosis.
Figures

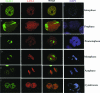
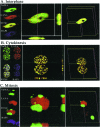


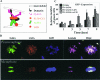
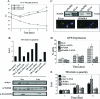
Similar articles
-
KSHV LANA--the master regulator of KSHV latency.Viruses. 2014 Dec 11;6(12):4961-98. doi: 10.3390/v6124961. Viruses. 2014. PMID: 25514370 Free PMC article. Review.
-
Kaposi's Sarcoma-Associated Herpesvirus LANA-Adjacent Regions with Distinct Functions in Episome Segregation or Maintenance.J Virol. 2019 Mar 5;93(6):e02158-18. doi: 10.1128/JVI.02158-18. Print 2019 Mar 15. J Virol. 2019. PMID: 30626680 Free PMC article.
-
Bub1 and CENP-F can contribute to Kaposi's sarcoma-associated herpesvirus genome persistence by targeting LANA to kinetochores.J Virol. 2010 Oct;84(19):9718-32. doi: 10.1128/JVI.00713-10. Epub 2010 Jul 21. J Virol. 2010. PMID: 20660191 Free PMC article.
-
Identification of Kaposi's sarcoma-associated herpesvirus LANA regions important for episome segregation, replication, and persistence.J Virol. 2013 Nov;87(22):12270-83. doi: 10.1128/JVI.01243-13. Epub 2013 Sep 4. J Virol. 2013. PMID: 24006437 Free PMC article.
-
The latency-associated nuclear antigen, a multifunctional protein central to Kaposi's sarcoma-associated herpesvirus latency.Future Microbiol. 2011 Dec;6(12):1399-413. doi: 10.2217/fmb.11.137. Future Microbiol. 2011. PMID: 22122438 Free PMC article. Review.
Cited by
-
Kaposi's sarcoma-associated herpesvirus-encoded LANA interacts with host KAP1 to facilitate establishment of viral latency.J Virol. 2014 Jul;88(13):7331-44. doi: 10.1128/JVI.00596-14. Epub 2014 Apr 16. J Virol. 2014. PMID: 24741090 Free PMC article.
-
KSHV Genome Replication and Maintenance.Front Microbiol. 2016 Feb 1;7:54. doi: 10.3389/fmicb.2016.00054. eCollection 2016. Front Microbiol. 2016. PMID: 26870016 Free PMC article. Review.
-
Kaposi's Sarcoma-Associated Herpesvirus Latency-Associated Nuclear Antigen: Replicating and Shielding Viral DNA during Viral Persistence.J Virol. 2017 Jun 26;91(14):e01083-16. doi: 10.1128/JVI.01083-16. Print 2017 Jul 15. J Virol. 2017. PMID: 28446671 Free PMC article. Review.
-
Comprehensive analysis of LANA interacting proteins essential for viral genome tethering and persistence.PLoS One. 2013 Sep 11;8(9):e74662. doi: 10.1371/journal.pone.0074662. eCollection 2013. PLoS One. 2013. PMID: 24040311 Free PMC article.
-
KSHV LANA--the master regulator of KSHV latency.Viruses. 2014 Dec 11;6(12):4961-98. doi: 10.3390/v6124961. Viruses. 2014. PMID: 25514370 Free PMC article. Review.
References
-
- An, F. Q., N. Compitello, E. Horwitz, M. Sramkoski, E. S. Knudsen, and R. Renne. 2004. The latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus modulates cellular gene expression and protects lymphoid cells from P16INK4A-induced cell cycle arrest. J. Biol. Chem. 2803862-3874. - PubMed
-
- Bajaj, B. G., S. C. Verma, K. Lan, M. A. Cotter, Z. L. Woodman, and E. S. Robertson. 2006. KSHV encoded LANA upregulates Pim-1 and is a substrate for its kinase activity. Virology 35118-28. - PubMed
-
- Ballestas, M. E., P. A. Chatis, and K. M. Kaye. 1999. Efficient persistence of extrachromosomal KSHV DNA mediated by latency-associated nuclear antigen. Science 284641-644. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- DEO017338/PHS HHS/United States
- CA108461/CA/NCI NIH HHS/United States
- R01 GM083988/GM/NIGMS NIH HHS/United States
- K99 CA126182/CA/NCI NIH HHS/United States
- AI067037/AI/NIAID NIH HHS/United States
- R01 AI067037/AI/NIAID NIH HHS/United States
- K99CA126182/CA/NCI NIH HHS/United States
- R01 CA108461/CA/NCI NIH HHS/United States
- CA072510/CA/NCI NIH HHS/United States
- DE01436/DE/NIDCR NIH HHS/United States
- R01 CA091792/CA/NCI NIH HHS/United States
- K99 CA126182-02/CA/NCI NIH HHS/United States
- CA091792/CA/NCI NIH HHS/United States
- K99 CA126182-01A1/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

