Stabilization of RelB requires multidomain interactions with p100/p52
- PMID: 18321863
- PMCID: PMC2431000
- DOI: 10.1074/jbc.M707898200
Stabilization of RelB requires multidomain interactions with p100/p52
Abstract
The NF-kappaB family member RelB has many properties not shared by other family members such as restricted subunit association and lack of regulation by the classical IkappaB proteins. We show that the protein level of RelB is significantly reduced in the absence of p100 and reduced even more when both p100 and p105 are absent. RelB stabilizes itself by directly interacting with p100, p105, and their processed products. However, RelB forms complexes with its partners using different interaction modes. Although the C-terminal ankyrin repeat domain of p105 is not involved in the RelB-p105 complex formation, all domains and flexible regions of each protein are engaged in the RelB-p100 complex. In several respects the RelB-p52 and RelB-p100 complexes are unique in the NF-kappaB family. The N-terminal domain of p100/p52 interacts with RelB but not RelA. The transcriptional activation domain of RelB, but not RelA, directly interacts with the processing region of p100. These unique protein-protein contacts explain why RelB prefers p52 as its dimeric partner for transcriptional activity and is retained in the cytoplasm as an inhibited complex by p100. This association-mediated stabilization of RelB implies a possible role for RelB in the processing of p100 into p52.
Figures
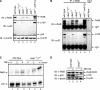


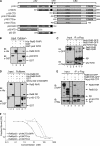


Similar articles
-
A structural basis for selective dimerization by NF-κB RelB.J Mol Biol. 2013 Jun 12;425(11):1934-1945. doi: 10.1016/j.jmb.2013.02.020. Epub 2013 Feb 26. J Mol Biol. 2013. PMID: 23485337
-
The NF-κB subunit RelB controls p100 processing by competing with the kinases NIK and IKK1 for binding to p100.Sci Signal. 2016 Sep 27;9(447):ra96. doi: 10.1126/scisignal.aad9413. Sci Signal. 2016. PMID: 27678221
-
The Nfkb1 and Nfkb2 proteins p105 and p100 function as the core of high-molecular-weight heterogeneous complexes.Mol Cell. 2009 Jun 12;34(5):591-602. doi: 10.1016/j.molcel.2009.04.033. Mol Cell. 2009. PMID: 19524538 Free PMC article.
-
RelB, a member of the Rel/NF-kappa B family of transcription factors.Braz J Med Biol Res. 1996 Jul;29(7):895-903. Braz J Med Biol Res. 1996. PMID: 9070378 Review.
-
Origin of the Functional Distinctiveness of NF-κB/p52.Front Cell Dev Biol. 2021 Nov 23;9:764164. doi: 10.3389/fcell.2021.764164. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34888310 Free PMC article. Review.
Cited by
-
B-cell CLL/lymphoma 10 (BCL10) is required for NF-kappaB production by both canonical and noncanonical pathways and for NF-kappaB-inducing kinase (NIK) phosphorylation.J Biol Chem. 2010 Jan 1;285(1):522-30. doi: 10.1074/jbc.M109.050815. Epub 2009 Nov 6. J Biol Chem. 2010. PMID: 19897484 Free PMC article.
-
IKK phosphorylates RelB to modulate its promoter specificity and promote fibroblast migration downstream of TNF receptors.Proc Natl Acad Sci U S A. 2014 Oct 14;111(41):14794-9. doi: 10.1073/pnas.1410124111. Epub 2014 Sep 29. Proc Natl Acad Sci U S A. 2014. PMID: 25267645 Free PMC article.
-
Non-canonical NF-κB signalling and ETS1/2 cooperatively drive C250T mutant TERT promoter activation.Nat Cell Biol. 2015 Oct;17(10):1327-38. doi: 10.1038/ncb3240. Epub 2015 Sep 21. Nat Cell Biol. 2015. PMID: 26389665 Free PMC article.
-
Stimulus-selective crosstalk via the NF-κB signaling system reinforces innate immune response to alleviate gut infection.Elife. 2015 Apr 23;4:e05648. doi: 10.7554/eLife.05648. Elife. 2015. PMID: 25905673 Free PMC article.
-
The NF-kappaB factor RelB and histone H3 lysine methyltransferase G9a directly interact to generate epigenetic silencing in endotoxin tolerance.J Biol Chem. 2009 Oct 9;284(41):27857-27865. doi: 10.1074/jbc.M109.000950. Epub 2009 Aug 18. J Biol Chem. 2009. PMID: 19690169 Free PMC article.
References
-
- Baldwin, A. S., Jr. (1996) Annu. Rev. Immunol. 14 649-683 - PubMed
-
- Ghosh, S., May, M. J., and Kopp, E. B. (1998) Annu. Rev. Immunol. 16 225-260 - PubMed
-
- Ghosh, S., and Karin, M. (2002) Cell 109 (Suppl.) S81-S96 - PubMed
-
- Karin, M., and Ben-Neriah, Y. (2000) Annu. Rev. Immunol. 18 621-663 - PubMed
-
- Solan, N. J., Miyoshi, H., Carmona, E. M., Bren, G. D., and Paya, C. V. (2002) J. Biol. Chem. 277 1405-1418 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

