Asn 362 in gp120 contributes to enhanced fusogenicity by CCR5-restricted HIV-1 envelope glycoprotein variants from patients with AIDS
- PMID: 18076768
- PMCID: PMC2225424
- DOI: 10.1186/1742-4690-4-89
Asn 362 in gp120 contributes to enhanced fusogenicity by CCR5-restricted HIV-1 envelope glycoprotein variants from patients with AIDS
Abstract
Background: CCR5-restricted (R5) human immunodeficiency virus type 1 (HIV-1) variants cause CD4+ T-cell loss in the majority of individuals who progress to AIDS, but mechanisms underlying the pathogenicity of R5 strains are poorly understood. To better understand envelope glycoprotein (Env) determinants contributing to pathogenicity of R5 viruses, we characterized 37 full-length R5 Envs from cross-sectional and longitudinal R5 viruses isolated from blood of patients with asymptomatic infection or AIDS, referred to as pre-AIDS (PA) and AIDS (A) R5 Envs, respectively.
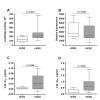
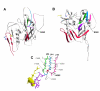
Results: Compared to PA-R5 Envs, A-R5 Envs had enhanced fusogenicity in quantitative cell-cell fusion assays, and reduced sensitivity to inhibition by the fusion inhibitor T-20. Sequence analysis identified the presence of Asn 362 (N362), a potential N-linked glycosylation site immediately N-terminal to CD4-binding site (CD4bs) residues in the C3 region of gp120, more frequently in A-R5 Envs than PA-R5 Envs. N362 was associated with enhanced fusogenicity, faster entry kinetics, and increased sensitivity of Env-pseudotyped reporter viruses to neutralization by the CD4bs-directed Env mAb IgG1b12. Mutagenesis studies showed N362 contributes to enhanced fusogenicity of most A-R5 Envs. Molecular models indicate N362 is located adjacent to the CD4 binding loop of gp120, and suggest N362 may enhance fusogenicity by promoting greater exposure of the CD4bs and/or stabilizing the CD4-bound Env structure.
Conclusion: Enhanced fusogenicity is a phenotype of the A-R5 Envs studied, which was associated with the presence of N362, enhanced HIV-1 entry kinetics and increased CD4bs exposure in gp120. N362 contributes to fusogenicity of R5 Envs in a strain dependent manner. Our studies suggest enhanced fusogenicity of A-R5 Envs may contribute to CD4+ T-cell loss in subjects who progress to AIDS whilst harbouring R5 HIV-1 variants. N362 may contribute to this effect in some individuals.
Figures



Similar articles
-
HIV-1 R5 Macrophage-Tropic Envelope Glycoprotein Trimers Bind CD4 with High Affinity, while the CD4 Binding Site on Non-macrophage-tropic, T-Tropic R5 Envelopes Is Occluded.J Virol. 2018 Jan 2;92(2):e00841-17. doi: 10.1128/JVI.00841-17. Print 2018 Jan 15. J Virol. 2018. PMID: 29118121 Free PMC article.
-
Enhanced CD4+ cellular apoptosis by CCR5-restricted HIV-1 envelope glycoprotein variants from patients with progressive HIV-1 infection.Virology. 2010 Jan 20;396(2):246-55. doi: 10.1016/j.virol.2009.10.029. Epub 2009 Nov 13. Virology. 2010. PMID: 19913863
-
CD4-binding site alterations in CCR5-using HIV-1 envelopes influencing gp120-CD4 interactions and fusogenicity.Virology. 2011 Feb 20;410(2):418-28. doi: 10.1016/j.virol.2010.12.010. Epub 2011 Jan 8. Virology. 2011. PMID: 21216423
-
HIV-1 envelope, integrins and co-receptor use in mucosal transmission of HIV.J Transl Med. 2011 Jan 27;9 Suppl 1(Suppl 1):S2. doi: 10.1186/1479-5876-9-S1-S2. J Transl Med. 2011. PMID: 21284901 Free PMC article. Review.
-
Variation of macrophage tropism among HIV-1 R5 envelopes in brain and other tissues.J Neuroimmune Pharmacol. 2007 Mar;2(1):32-41. doi: 10.1007/s11481-006-9042-2. Epub 2006 Nov 7. J Neuroimmune Pharmacol. 2007. PMID: 18040824 Review.
Cited by
-
A quantitative affinity-profiling system that reveals distinct CD4/CCR5 usage patterns among human immunodeficiency virus type 1 and simian immunodeficiency virus strains.J Virol. 2009 Nov;83(21):11016-26. doi: 10.1128/JVI.01242-09. Epub 2009 Aug 19. J Virol. 2009. PMID: 19692480 Free PMC article.
-
Determinants flanking the CD4 binding loop modulate macrophage tropism of human immunodeficiency virus type 1 R5 envelopes.J Virol. 2009 Mar;83(6):2575-83. doi: 10.1128/JVI.02133-08. Epub 2009 Jan 7. J Virol. 2009. PMID: 19129457 Free PMC article.
-
Alternative coreceptor requirements for efficient CCR5- and CXCR4-mediated HIV-1 entry into macrophages.J Virol. 2011 Oct;85(20):10699-709. doi: 10.1128/JVI.05510-11. Epub 2011 Aug 10. J Virol. 2011. PMID: 21835796 Free PMC article.
-
Single genome analysis reveals genetic characteristics of Neuroadaptation across HIV-1 envelope.Retrovirology. 2014 Aug 15;11:65. doi: 10.1186/s12977-014-0065-0. Retrovirology. 2014. PMID: 25125210 Free PMC article.
-
The HIV-1 env protein: a coat of many colors.Curr HIV/AIDS Rep. 2012 Mar;9(1):52-63. doi: 10.1007/s11904-011-0107-3. Curr HIV/AIDS Rep. 2012. PMID: 22237899 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

