Promiscuous mutations activate the noncanonical NF-kappaB pathway in multiple myeloma
- PMID: 17692805
- PMCID: PMC2083698
- DOI: 10.1016/j.ccr.2007.07.003
Promiscuous mutations activate the noncanonical NF-kappaB pathway in multiple myeloma
Abstract
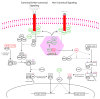
Activation of NF-kappaB has been noted in many tumor types, however only rarely has this been linked to an underlying genetic mutation. An integrated analysis of high-density oligonucleotide array CGH and gene expression profiling data from 155 multiple myeloma samples identified a promiscuous array of abnormalities contributing to the dysregulation of NF-kappaB in approximately 20% of patients. We report mutations in ten genes causing the inactivation of TRAF2, TRAF3, CYLD, cIAP1/cIAP2 and activation of NFKB1, NFKB2, CD40, LTBR, TACI, and NIK that result primarily in constitutive activation of the noncanonical NF-kappaB pathway, with the single most common abnormality being inactivation of TRAF3. These results highlight the critical importance of the NF-kappaB pathway in the pathogenesis of multiple myeloma.
Figures



Similar articles
-
Classical and/or alternative NF-kappaB pathway activation in multiple myeloma.Blood. 2010 Apr 29;115(17):3541-52. doi: 10.1182/blood-2009-09-243535. Epub 2010 Jan 6. Blood. 2010. PMID: 20053756 Free PMC article.
-
Frequent engagement of the classical and alternative NF-kappaB pathways by diverse genetic abnormalities in multiple myeloma.Cancer Cell. 2007 Aug;12(2):115-30. doi: 10.1016/j.ccr.2007.07.004. Cancer Cell. 2007. PMID: 17692804 Free PMC article.
-
TACI induces cIAP1-mediated ubiquitination of NIK by TRAF2 and TANK to limit non-canonical NF-kappaB signaling.J Recept Signal Transduct Res. 2010 Apr;30(2):121-32. doi: 10.3109/10799891003634509. J Recept Signal Transduct Res. 2010. PMID: 20184394
-
SCF-mediated degradation of p100 (NF-κB2): mechanisms and relevance in multiple myeloma.Sci Signal. 2012 Dec 4;5(253):pt14. doi: 10.1126/scisignal.2003408. Sci Signal. 2012. PMID: 23211527 Free PMC article. Review.
-
Targeting signaling factors for degradation, an emerging mechanism for TRAF functions.Immunol Rev. 2015 Jul;266(1):56-71. doi: 10.1111/imr.12311. Immunol Rev. 2015. PMID: 26085207 Free PMC article. Review.
Cited by
-
Downregulation of TRAF2 mediates NIK-induced pancreatic cancer cell proliferation and tumorigenicity.PLoS One. 2013;8(1):e53676. doi: 10.1371/journal.pone.0053676. Epub 2013 Jan 3. PLoS One. 2013. PMID: 23301098 Free PMC article.
-
An oncogenic role for alternative NF-κB signaling in DLBCL revealed upon deregulated BCL6 expression.Cell Rep. 2015 May 5;11(5):715-26. doi: 10.1016/j.celrep.2015.03.059. Epub 2015 Apr 23. Cell Rep. 2015. PMID: 25921526 Free PMC article.
-
Osteoclasts-Key Players in Skeletal Health and Disease.Microbiol Spectr. 2016 Jun;4(3):10.1128/microbiolspec.MCHD-0011-2015. doi: 10.1128/microbiolspec.MCHD-0011-2015. Microbiol Spectr. 2016. PMID: 27337470 Free PMC article. Review.
-
Inactivation of CYLD in intestinal epithelial cells exacerbates colitis-associated colorectal carcinogenesis - a short report.Cell Oncol (Dordr). 2016 Jun;39(3):287-93. doi: 10.1007/s13402-016-0279-3. Epub 2016 Apr 4. Cell Oncol (Dordr). 2016. PMID: 27042826
-
APRIL Drives a Coordinated but Diverse Response as a Foundation for Plasma Cell Longevity.J Immunol. 2022 Sep 1;209(5):926-937. doi: 10.4049/jimmunol.2100623. Epub 2022 Aug 5. J Immunol. 2022. PMID: 36130130 Free PMC article.
References
-
- Annunziata CM, Davis R, Gabrea A, Kuehl M, Staudt LM. NF-kappaB-inducing kinase activates NF-kappaB signaling in multiple myeloma. Proc Amer Assoc Cancer Res. 2006;47 [Abstract #LB-124]
Publication types
MeSH terms
Substances
Grants and funding
- R01 CA083724/CA/NCI NIH HHS/United States
- R01 CA083724-07/CA/NCI NIH HHS/United States
- P01 CA62242/CA/NCI NIH HHS/United States
- R01 AG020686-01A1/AG/NIA NIH HHS/United States
- R01 AG020686/AG/NIA NIH HHS/United States
- P50 CA100707-01/CA/NCI NIH HHS/United States
- R01 AG020686-04/AG/NIA NIH HHS/United States
- R01 AG020686-05/AG/NIA NIH HHS/United States
- R01 AG020686-03/AG/NIA NIH HHS/United States
- P50 CA100707-010005/CA/NCI NIH HHS/United States
- P01 CA062242/CA/NCI NIH HHS/United States
- P50 CA100707-010004/CA/NCI NIH HHS/United States
- P50 CA100707/CA/NCI NIH HHS/United States
- R01 CA83724-01/CA/NCI NIH HHS/United States
- R01 AG020686-02/AG/NIA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

