Positional stability of single double-strand breaks in mammalian cells
- PMID: 17486118
- PMCID: PMC2442898
- DOI: 10.1038/ncb1591
Positional stability of single double-strand breaks in mammalian cells
Abstract
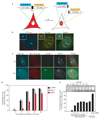
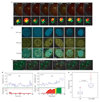
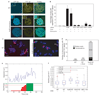
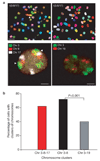
Formation of cancerous translocations requires the illegitimate joining of chromosomes containing double-strand breaks (DSBs). It is unknown how broken chromosome ends find their translocation partners within the cell nucleus. Here, we have visualized and quantitatively analysed the dynamics of single DSBs in living mammalian cells. We demonstrate that broken ends are positionally stable and unable to roam the cell nucleus. Immobilization of broken chromosome ends requires the DNA-end binding protein Ku80, but is independent of DNA repair factors, H2AX, the MRN complex and the cohesion complex. DSBs preferentially undergo translocations with neighbouring chromosomes and loss of local positional constraint correlates with elevated genomic instability. These results support a contact-first model in which chromosome translocations predominantly form among spatially proximal DSBs.
Figures
Similar articles
-
Nonhomologous DNA end joining and chromosome aberrations in human embryonic lung fibroblasts treated with environmental pollutants.Mutat Res. 2014 May-Jun;763-764:28-38. doi: 10.1016/j.mrfmmm.2014.03.006. Epub 2014 Mar 30. Mutat Res. 2014. PMID: 24694657
-
Phosphorylation of Ku dictates DNA double-strand break (DSB) repair pathway choice in S phase.Nucleic Acids Res. 2016 Feb 29;44(4):1732-45. doi: 10.1093/nar/gkv1499. Epub 2015 Dec 27. Nucleic Acids Res. 2016. PMID: 26712563 Free PMC article.
-
Binding of double-strand breaks in DNA by human Rad52 protein.Nature. 1999 Apr 22;398(6729):728-31. doi: 10.1038/19560. Nature. 1999. PMID: 10227297
-
Telomere biology: integrating chromosomal end protection with DNA damage response.Chromosoma. 2005 Sep;114(4):275-85. doi: 10.1007/s00412-005-0338-4. Epub 2005 Oct 15. Chromosoma. 2005. PMID: 15843951 Review.
-
Dynamics of double strand breaks and chromosomal translocations.Mol Cancer. 2014 Nov 18;13:249. doi: 10.1186/1476-4598-13-249. Mol Cancer. 2014. PMID: 25404525 Free PMC article. Review.
Cited by
-
Tissue microarray analyses of the essential DNA repair factors ATM, DNA-PKcs and Ku80 in head and neck squamous cell carcinoma.Radiat Oncol. 2024 Oct 30;19(1):150. doi: 10.1186/s13014-024-02541-3. Radiat Oncol. 2024. PMID: 39478631 Free PMC article.
-
DNA double-strand break-capturing nuclear envelope tubules drive DNA repair.Nat Struct Mol Biol. 2024 Sep;31(9):1319-1330. doi: 10.1038/s41594-024-01286-7. Epub 2024 Apr 17. Nat Struct Mol Biol. 2024. PMID: 38632359
-
Multi-Scale Imaging of the Dynamic Organization of Chromatin.Int J Mol Sci. 2023 Nov 4;24(21):15975. doi: 10.3390/ijms242115975. Int J Mol Sci. 2023. PMID: 37958958 Free PMC article. Review.
-
DNA double-strand break end synapsis by DNA loop extrusion.Nat Commun. 2023 Apr 6;14(1):1913. doi: 10.1038/s41467-023-37583-w. Nat Commun. 2023. PMID: 37024496 Free PMC article.
-
Let's not take DNA breaks for granted. The importance of direct detection of DNA breaks for the successful development of DDR inhibitors.Front Cell Dev Biol. 2023 Mar 9;11:1118716. doi: 10.3389/fcell.2023.1118716. eCollection 2023. Front Cell Dev Biol. 2023. PMID: 36968210 Free PMC article. Review.
References
-
- Kanaar R, Hoeijmakers JH, van Gent DC. Molecular mechanisms of DNA double strand break repair. Trends Cell Biol. 1998;8:483–489. - PubMed
-
- Khanna KK, Jackson SP. DNA double-strand breaks: signaling, repair and the cancer connection. Nature Genet. 2001;27:247–254. - PubMed
-
- Nikiforova MN, et al. Proximity of chromosomal loci that participate in radiation-induced rearrangements in human cells. Science. 2000;290:138–141. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

