Origin and evolution of human microRNAs from transposable elements
- PMID: 17435244
- PMCID: PMC1894593
- DOI: 10.1534/genetics.107.072553
Origin and evolution of human microRNAs from transposable elements
Abstract
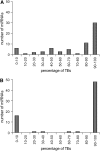
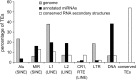
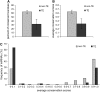
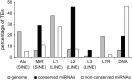
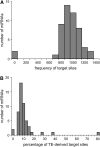
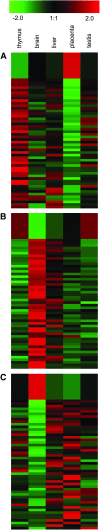
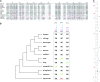
We sought to evaluate the extent of the contribution of transposable elements (TEs) to human microRNA (miRNA) genes along with the evolutionary dynamics of TE-derived human miRNAs. We found 55 experimentally characterized human miRNA genes that are derived from TEs, and these TE-derived miRNAs have the potential to regulate thousands of human genes. Sequence comparisons revealed that TE-derived human miRNAs are less conserved, on average, than non-TE-derived miRNAs. However, there are 18 TE-derived miRNAs that are relatively conserved, and 14 of these are related to the ancient L2 and MIR families. Comparison of miRNA vs. mRNA expression patterns for TE-derived miRNAs and their putative target genes showed numerous cases of anti-correlated expression that are consistent with regulation via mRNA degradation. In addition to the known human miRNAs that we show to be derived from TE sequences, we predict an additional 85 novel TE-derived miRNA genes. TE sequences are typically disregarded in genomic surveys for miRNA genes and target sites; this is a mistake. Our results indicate that TEs provide a natural mechanism for the origination miRNAs that can contribute to regulatory divergence between species as well as a rich source for the discovery of as yet unknown miRNA genes.
Figures
Similar articles
-
Functional microRNAs and target sites are created by lineage-specific transposition.Hum Mol Genet. 2014 Apr 1;23(7):1783-93. doi: 10.1093/hmg/ddt569. Epub 2013 Nov 13. Hum Mol Genet. 2014. PMID: 24234653 Free PMC article.
-
The Role of Transposable Elements in the Origin and Evolution of MicroRNAs in Human.PLoS One. 2015 Jun 26;10(6):e0131365. doi: 10.1371/journal.pone.0131365. eCollection 2015. PLoS One. 2015. PMID: 26115450 Free PMC article.
-
Domestication of transposable elements into MicroRNA genes in plants.PLoS One. 2011 May 3;6(5):e19212. doi: 10.1371/journal.pone.0019212. PLoS One. 2011. PMID: 21559273 Free PMC article.
-
Origin of a substantial fraction of human regulatory sequences from transposable elements.Trends Genet. 2003 Feb;19(2):68-72. doi: 10.1016/s0168-9525(02)00006-9. Trends Genet. 2003. PMID: 12547512 Review.
-
Bioinformatics Analysis of Evolution and Human Disease Related Transposable Element-Derived microRNAs.Life (Basel). 2020 Jun 25;10(6):95. doi: 10.3390/life10060095. Life (Basel). 2020. PMID: 32630504 Free PMC article. Review.
Cited by
-
Nothing in Evolution Makes Sense Except in the Light of Genomics: Read-Write Genome Evolution as an Active Biological Process.Biology (Basel). 2016 Jun 8;5(2):27. doi: 10.3390/biology5020027. Biology (Basel). 2016. PMID: 27338490 Free PMC article. Review.
-
Losing identity: structural diversity of transposable elements belonging to different classes in the genome of Anopheles gambiae.BMC Genomics. 2012 Jun 22;13:272. doi: 10.1186/1471-2164-13-272. BMC Genomics. 2012. PMID: 22726298 Free PMC article.
-
Retrotransposons as Drivers of Mammalian Brain Evolution.Life (Basel). 2021 Apr 22;11(5):376. doi: 10.3390/life11050376. Life (Basel). 2021. PMID: 33922141 Free PMC article. Review.
-
Regulators of gene expression as biomarkers for prostate cancer.Am J Cancer Res. 2012;2(6):620-57. Epub 2012 Nov 20. Am J Cancer Res. 2012. PMID: 23226612 Free PMC article.
-
Generation of de novo miRNAs from template switching during DNA replication.Proc Natl Acad Sci U S A. 2023 Dec 5;120(49):e2310752120. doi: 10.1073/pnas.2310752120. Epub 2023 Nov 29. Proc Natl Acad Sci U S A. 2023. PMID: 38019864 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

