Methylation-controlled J protein promotes c-Jun degradation to prevent ABCB1 transporter expression
- PMID: 17283040
- PMCID: PMC1899938
- DOI: 10.1128/MCB.01804-06
Methylation-controlled J protein promotes c-Jun degradation to prevent ABCB1 transporter expression
Abstract
Methylation-controlled J protein (MCJ) is a newly identified member of the DnaJ family of cochaperones. Hypermethylation-mediated transcriptional silencing of the MCJ gene has been associated with increased chemotherapeutic resistance in ovarian cancer. However, the biology and function of MCJ remain unknown. Here we show that MCJ is a type II transmembrane cochaperone localized in the Golgi network and present only in vertebrates. MCJ is expressed in drug-sensitive breast cancer cells but not in multidrug-resistant cells. The inhibition of MCJ expression increases resistance to specific drugs by inducing expression of the ABCB1 drug transporter that prevents intracellular drug accumulation. The induction of ABCB1 gene expression is mediated by increased levels of c-Jun due to an impaired degradation of this transcription factor in the absence of MCJ. Thus, MCJ is required in these cells to prevent c-Jun-mediated expression of ABCB1 and maintain drug response.
Figures


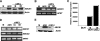

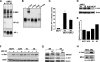
Similar articles
-
Loss of expression of a new member of the DNAJ protein family confers resistance to chemotherapeutic agents used in the treatment of ovarian cancer.Cancer Res. 2001 May 15;61(10):4258-65. Cancer Res. 2001. PMID: 11358853
-
Demethylation of the MCJ gene in stage III/IV epithelial ovarian cancer and response to chemotherapy.Gynecol Oncol. 2005 Jun;97(3):898-903. doi: 10.1016/j.ygyno.2005.03.023. Gynecol Oncol. 2005. PMID: 15894365
-
ABCB1 over-expression and drug-efflux in acute lymphoblastic leukemia cell lines with t(17;19) and E2A-HLF expression.Pediatr Blood Cancer. 2006 Nov;47(6):757-64. doi: 10.1002/pbc.20635. Pediatr Blood Cancer. 2006. PMID: 16206189
-
ABCB1 genotype and PGP expression, function and therapeutic drug response: a critical review and recommendations for future research.Pharmacogenomics J. 2007 Jun;7(3):154-79. doi: 10.1038/sj.tpj.6500413. Epub 2006 Sep 12. Pharmacogenomics J. 2007. PMID: 16969364 Review.
-
ABCB1 pharmacogenetics: progress, pitfalls, and promise.Clin Pharmacol Ther. 2007 Feb;81(2):265-9. doi: 10.1038/sj.clpt.6100052. Clin Pharmacol Ther. 2007. PMID: 17259950 Review.
Cited by
-
MCJ/DnaJC15, an endogenous mitochondrial repressor of the respiratory chain that controls metabolic alterations.Mol Cell Biol. 2013 Jun;33(11):2302-14. doi: 10.1128/MCB.00189-13. Epub 2013 Mar 25. Mol Cell Biol. 2013. PMID: 23530063 Free PMC article.
-
Integration and bioinformatics analysis of DNA-methylated genes associated with drug resistance in ovarian cancer.Oncol Lett. 2016 Jul;12(1):157-166. doi: 10.3892/ol.2016.4608. Epub 2016 May 18. Oncol Lett. 2016. PMID: 27347118 Free PMC article.
-
Meta-analysis of gene signatures and key pathways indicates suppression of JNK pathway as a regulator of chemo-resistance in AML.Sci Rep. 2021 Jun 14;11(1):12485. doi: 10.1038/s41598-021-91864-2. Sci Rep. 2021. PMID: 34127725 Free PMC article.
-
Nuclear localization of p38 MAPK in response to DNA damage.Int J Biol Sci. 2009 Jun 16;5(5):428-37. doi: 10.7150/ijbs.5.428. Int J Biol Sci. 2009. PMID: 19564926 Free PMC article.
-
Regulation of oxidative stress by methylation-controlled J protein controls macrophage responses to inflammatory insults.J Infect Dis. 2015 Jan 1;211(1):135-45. doi: 10.1093/infdis/jiu389. Epub 2014 Jul 14. J Infect Dis. 2015. PMID: 25028693 Free PMC article.
References
-
- Alley, M., D. Scudiero, A. Monks, M. Hursey, M. Czerwinski, D. Fine, B. Abbott, J. Mayo, R. Shoemaker, and M. Boyd. 1988. Feasibility of drug screening with panels of human tumor cell lines using a microculture tetrazolium assay. Cancer Res. 48:589-601. - PubMed
-
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. - PubMed
-
- Angel, P., M. Imagawa, R. Chiu, B. Stein, R. J. Imbra, H. J. Rahmsdorf, C. Jonat, P. Herrlich, and M. Karin. 1987. Phorbol ester-inducible genes contain a common cis element recognized by a TPA-modulated trans-acting factor. Cell 49:729-739. - PubMed
-
- Belhoussine, R., H. Morjani, S. Sharonov, D. Ploton, and M. Manfait. 1999. Characterization of intracellular pH gradients in human multidrug-resistant tumor cells by means of scanning microspectrofluorometry and dual-emission-ratio probes. Int. J. Cancer 81:81-89. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
