Downregulation of gamma interferon receptor 1 by Kaposi's sarcoma-associated herpesvirus K3 and K5
- PMID: 17166914
- PMCID: PMC1865953
- DOI: 10.1128/JVI.01961-06
Downregulation of gamma interferon receptor 1 by Kaposi's sarcoma-associated herpesvirus K3 and K5
Abstract
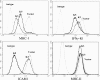
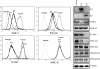
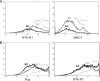
Upon viral infection, the major defense mounted by the host immune system is activation of the interferon (IFN)-mediated antiviral pathway. In order to complete their life cycles, viruses must modulate the host IFN-mediated immune response. The K3 and K5 proteins of a human tumor-inducing herpesvirus, Kaposi's sarcoma-associated herpesvirus (KSHV), have been shown to downregulate the surface expression of host immune modulatory receptors by increasing their endocytosis rates, which leads to suppression of cell-mediated immunity. In this report, we demonstrate that K3 and K5 both specifically target gamma interferon receptor 1 (IFN-gammaR1) and induce its ubiquitination, endocytosis, and degradation, resulting in downregulation of IFN-gammaR1 surface expression and, thereby, inhibition of IFN-gamma action. Mutational analysis indicated that K5 appeared to downregulate IFN-gammaR1 more strongly than K3 and that the amino-terminal ring finger motif and the carboxyl-terminal region of K5 were necessary for IFN-gammaR1 downregulation. These results suggest that KSHV K3 and K5 suppress both cytokine-mediated and cell-mediated immunity, which ensures efficient viral avoidance of host immune controls.
Figures




Similar articles
-
Inhibition of interferon regulatory factor 7 (IRF7)-mediated interferon signal transduction by the Kaposi's sarcoma-associated herpesvirus viral IRF homolog vIRF3.J Virol. 2007 Aug;81(15):8282-92. doi: 10.1128/JVI.00235-07. Epub 2007 May 23. J Virol. 2007. PMID: 17522209 Free PMC article.
-
Human airway macrophages are metabolically reprogrammed by IFN-γ resulting in glycolysis-dependent functional plasticity.Elife. 2024 Dec 2;13:RP98449. doi: 10.7554/eLife.98449. Elife. 2024. PMID: 39620891 Free PMC article.
-
Depressing time: Waiting, melancholia, and the psychoanalytic practice of care.In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. PMID: 36137063 Free Books & Documents. Review.
-
Modulation of Lymphotoxin β Surface Expression by Kaposi's Sarcoma-Associated Herpesvirus K3 Through Glycosylation Interference.J Med Virol. 2025 Jan;97(1):e70179. doi: 10.1002/jmv.70179. J Med Virol. 2025. PMID: 39831393 Free PMC article.
-
Trends in Surgical and Nonsurgical Aesthetic Procedures: A 14-Year Analysis of the International Society of Aesthetic Plastic Surgery-ISAPS.Aesthetic Plast Surg. 2024 Oct;48(20):4217-4227. doi: 10.1007/s00266-024-04260-2. Epub 2024 Aug 5. Aesthetic Plast Surg. 2024. PMID: 39103642 Review.
Cited by
-
Interferon gamma receptor: the beginning of the journey.Front Immunol. 2013 Sep 3;4:267. doi: 10.3389/fimmu.2013.00267. Front Immunol. 2013. PMID: 24027571 Free PMC article. Review.
-
Kaposi's sarcoma-associated herpesvirus K3 and K5 ubiquitin E3 ligases have stage-specific immune evasion roles during lytic replication.J Virol. 2014 Aug;88(16):9335-49. doi: 10.1128/JVI.00873-14. Epub 2014 Jun 4. J Virol. 2014. PMID: 24899205 Free PMC article.
-
A Review of Functional Motifs Utilized by Viruses.Proteomes. 2016 Jan 21;4(1):3. doi: 10.3390/proteomes4010003. Proteomes. 2016. PMID: 28248213 Free PMC article. Review.
-
Ubiquitin-mediated regulation of CD86 protein expression by the ubiquitin ligase membrane-associated RING-CH-1 (MARCH1).J Biol Chem. 2011 Oct 28;286(43):37168-80. doi: 10.1074/jbc.M110.204040. Epub 2011 Sep 6. J Biol Chem. 2011. PMID: 21896490 Free PMC article.
-
KSHV-Mediated Angiogenesis in Tumor Progression.Viruses. 2016 Jul 20;8(7):198. doi: 10.3390/v8070198. Viruses. 2016. PMID: 27447661 Free PMC article. Review.
References
-
- Bach, E. A., M. Aguet, and R. D. Schreiber. 1997. The IFN gamma receptor: a paradigm for cytokine receptor signaling. Annu. Rev. Immunol. 15:563-591. - PubMed
-
- Bandres, J. C., A. S. Shaw, and L. Ratner. 1995. HIV-1 Nef protein downregulation of CD4 surface expression: relevance of the lck binding domain of CD4. Virology 207:338-341. - PubMed
-
- Barnes, B., B. Lubyova, and P. M. Pitha. 2002. On the role of IRF in host defense. J. Interferon Cytokine Res. 22:59-71. - PubMed
-
- Billiau, A. 1996. Interferon-gamma: biology and role in pathogenesis. Adv. Immunol. 62:61-130. - PubMed
-
- Boname, J. M., and P. G. Stevenson. 2001. MHC class I ubiquitination by a viral PHD/LAP finger protein. Immunity 15:627-636. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- CA 102535/CA/NCI NIH HHS/United States
- RR 00168/RR/NCRR NIH HHS/United States
- CA 91819/CA/NCI NIH HHS/United States
- CA 082057/CA/NCI NIH HHS/United States
- R01 CA082057/CA/NCI NIH HHS/United States
- P51 RR000168/RR/NCRR NIH HHS/United States
- K26 RR000168/RR/NCRR NIH HHS/United States
- R01 CA091819/CA/NCI NIH HHS/United States
- CA 31363/CA/NCI NIH HHS/United States
- R21 CA102535/CA/NCI NIH HHS/United States
- R01 CA086038/CA/NCI NIH HHS/United States
- CA 86038/CA/NCI NIH HHS/United States
- R01 CA031363/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

