Genes of the antioxidant system of the honey bee: annotation and phylogeny
- PMID: 17069640
- PMCID: PMC1847502
- DOI: 10.1111/j.1365-2583.2006.00695.x
Genes of the antioxidant system of the honey bee: annotation and phylogeny
Abstract
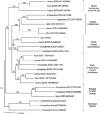
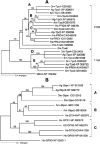
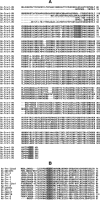
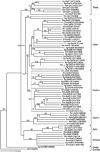
Antioxidant enzymes perform a variety of vital functions including the reduction of life-shortening oxidative damage. We used the honey bee genome sequence to identify the major components of the honey bee antioxidant system. A comparative analysis of honey bee with Drosophila melanogaster and Anopheles gambiae shows that although the basic components of the antioxidant system are conserved, there are important species differences in the number of paralogs. These include the duplication of thioredoxin reductase and the expansion of the thioredoxin family in fly; lack of expansion of the Theta, Delta and Omega GST classes in bee and no expansion of the Sigma class in dipteran species. The differential expansion of antioxidant gene families among honey bees and dipteran species might reflect the marked differences in life history and ecological niches between social and solitary species.
Figures


Similar articles
-
A putative glutathione peroxidase of Drosophila encodes a thioredoxin peroxidase that provides resistance against oxidative stress but fails to complement a lack of catalase activity.Biol Chem. 2003 Mar;384(3):463-72. doi: 10.1515/BC.2003.052. Biol Chem. 2003. PMID: 12715897
-
The nicotinic acetylcholine receptor gene family of the honey bee, Apis mellifera.Genome Res. 2006 Nov;16(11):1422-30. doi: 10.1101/gr.4549206. Epub 2006 Oct 25. Genome Res. 2006. PMID: 17065616 Free PMC article.
-
Function and evolution of a gene family encoding odorant binding-like proteins in a social insect, the honey bee (Apis mellifera).Genome Res. 2006 Nov;16(11):1404-13. doi: 10.1101/gr.5075706. Epub 2006 Oct 25. Genome Res. 2006. PMID: 17065610 Free PMC article.
-
The thioredoxin antioxidant system.Free Radic Biol Med. 2014 Jan;66:75-87. doi: 10.1016/j.freeradbiomed.2013.07.036. Epub 2013 Jul 27. Free Radic Biol Med. 2014. PMID: 23899494 Review.
-
Redox and antioxidant systems of the malaria parasite Plasmodium falciparum.Mol Microbiol. 2004 Sep;53(5):1291-305. doi: 10.1111/j.1365-2958.2004.04257.x. Mol Microbiol. 2004. PMID: 15387810 Review.
Cited by
-
Identification, genomic organization, and oxidative stress response of a sigma class glutathione S-transferase gene (AccGSTS1) in the honey bee, Apis cerana cerana.Cell Stress Chaperones. 2013 Jul;18(4):415-26. doi: 10.1007/s12192-012-0394-7. Epub 2012 Dec 20. Cell Stress Chaperones. 2013. PMID: 23250585 Free PMC article.
-
Validation of quantitative real-time PCR reference genes and spatial expression profiles of detoxication-related genes under pesticide induction in honey bee, Apis mellifera.PLoS One. 2022 Nov 10;17(11):e0277455. doi: 10.1371/journal.pone.0277455. eCollection 2022. PLoS One. 2022. PMID: 36355804 Free PMC article.
-
Knockdown of selenocysteine-specific elongation factor in Amblyomma maculatum alters the pathogen burden of Rickettsia parkeri with epigenetic control by the Sin3 histone deacetylase corepressor complex.PLoS One. 2013 Nov 25;8(11):e82012. doi: 10.1371/journal.pone.0082012. eCollection 2013. PLoS One. 2013. PMID: 24282621 Free PMC article.
-
Reproductive Cessation and Post-Reproductive Lifespan in Honeybee Workers.Biology (Basel). 2024 Apr 24;13(5):287. doi: 10.3390/biology13050287. Biology (Basel). 2024. PMID: 38785769 Free PMC article.
-
Changes in the Honeybee Antioxidant System after 12 h of Exposure to Electromagnetic Field Frequency of 50 Hz and Variable Intensity.Insects. 2020 Oct 18;11(10):713. doi: 10.3390/insects11100713. Insects. 2020. PMID: 33081029 Free PMC article.
References
-
- Arner ES, Holmgren A. Physiological functions of thioredoxin and thioredoxin reductase. Eur J Biochem. 2000;267:6102–6109. - PubMed
-
- Bannai H, Tamada Y, Maruyama O, Nakai K, Miyano S. Extensive feature detection of N-terminal protein sorting signals. Bioinformatics. 2002;18:298–305. - PubMed
-
- Bauer H, Kanzok SM, Schirmer RH. Thioredoxin-2 but not thioredoxin-1 is a substrate of thioredoxin peroxidase-1 from Drosophila melanogaster: isolation and characterization of a second thioredoxin in Drosophila melanogaster and evidence for distinct biological functions of Trx-1 and Trx-2. J Biol Chem. 2002;277:17457–17463. - PubMed
-
- Bauer H, Gromer S, Urbani A, Schnolzer M, Schirmer RH, Muller HM. Thioredoxin reductase from the malaria mosquito Anopheles gambiae. Eur J Biochem. 2003;270:4272–4281. - PubMed
-
- Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J Mol Biol. 2004;340:783–795. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

