Epstein-Barr virus nuclear antigen 2 trans-activates the cellular antiapoptotic bfl-1 gene by a CBF1/RBPJ kappa-dependent pathway
- PMID: 16873269
- PMCID: PMC1563820
- DOI: 10.1128/JVI.00278-06
Epstein-Barr virus nuclear antigen 2 trans-activates the cellular antiapoptotic bfl-1 gene by a CBF1/RBPJ kappa-dependent pathway
Abstract
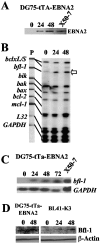
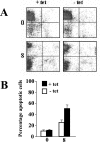
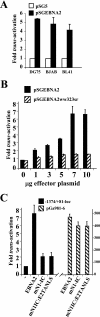
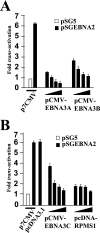
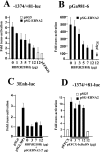
The human herpesvirus Epstein-Barr virus (EBV) establishes latency and promotes the long-term survival of its host B cell by targeting the molecular machinery controlling cell fate decisions. The cellular antiapoptotic bfl-1 gene confers protection from apoptosis under conditions of growth factor deprivation when expressed ectopically in an EBV-negative Burkitt's lymphoma-derived cell line (B. D'Souza, M. Rowe, and D. Walls, J. Virol. 74:6652-6658, 2000), and the EBV latent membrane protein 1 (LMP1) and its cellular functional homologue CD40 can both drive bfl-1 via an NF-kappaB-dependent enhancer element in the bfl-1 promoter (B. N. D'Souza, L. C. Edelstein, P. M. Pegman, S. M. Smith, S. T. Loughran, A. Clarke, A. Mehl, M. Rowe, C. Gélinas, and D. Walls, J. Virol. 78:1800-1816, 2004). Here we show that the EBV nuclear antigen 2 (EBNA2) also upregulates bfl-1. EBNA2 trans-activation of bfl-1 requires CBF1 (or RBP-J kappa), a nuclear component of the Notch signaling pathway, and there is an essential role for a core consensus CBF1-binding site on the bfl-1 promoter. trans-activation is dependent on the EBNA2-CBF1 interaction, is modulated by other EBV gene products known to interact with the CBF1 corepressor complex, and does not involve activation of NF-kappaB. bfl-1 expression is induced and maintained at high levels by the EBV growth program in a lymphoblastoid cell line, and withdrawal of either EBNA2 or LMP1 does not lead to a reduction in bfl-1 mRNA levels in this context, whereas the simultaneous loss of both EBV proteins results in a major decrease in bfl-1 expression. These findings are relevant to our understanding of EBV persistence, its role in malignant disease, and the B-cell developmental process.
Figures
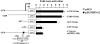


Similar articles
-
Nuclear factor kappa B-dependent activation of the antiapoptotic bfl-1 gene by the Epstein-Barr virus latent membrane protein 1 and activated CD40 receptor.J Virol. 2004 Feb;78(4):1800-16. doi: 10.1128/jvi.78.4.1800-1816.2004. J Virol. 2004. PMID: 14747545 Free PMC article.
-
The bfl-1 gene is transcriptionally upregulated by the Epstein-Barr virus LMP1, and its expression promotes the survival of a Burkitt's lymphoma cell line.J Virol. 2000 Jul;74(14):6652-8. doi: 10.1128/jvi.74.14.6652-6658.2000. J Virol. 2000. PMID: 10864681 Free PMC article.
-
The NP9 protein encoded by the human endogenous retrovirus HERV-K(HML-2) negatively regulates gene activation of the Epstein-Barr virus nuclear antigen 2 (EBNA2).Int J Cancer. 2011 Sep 1;129(5):1105-15. doi: 10.1002/ijc.25760. Epub 2011 Jan 12. Int J Cancer. 2011. PMID: 21710493
-
EBNA2 and Notch signalling in Epstein-Barr virus mediated immortalization of B lymphocytes.Semin Cancer Biol. 2001 Dec;11(6):423-34. doi: 10.1006/scbi.2001.0409. Semin Cancer Biol. 2001. PMID: 11669604 Review.
-
Both Epstein-Barr viral nuclear antigen 2 (EBNA2) and activated Notch1 transactivate genes by interacting with the cellular protein RBP-J kappa.Immunobiology. 1997 Dec;198(1-3):299-306. doi: 10.1016/s0171-2985(97)80050-2. Immunobiology. 1997. PMID: 9442401 Review.
Cited by
-
Repression of the proapoptotic cellular BIK/NBK gene by Epstein-Barr virus antagonizes transforming growth factor β1-induced B-cell apoptosis.J Virol. 2014 May;88(9):5001-13. doi: 10.1128/JVI.03642-13. Epub 2014 Feb 19. J Virol. 2014. PMID: 24554662 Free PMC article.
-
Epstein-Barr virus interactions with the Bcl-2 protein family and apoptosis in human tumor cells.J Zhejiang Univ Sci B. 2013 Jan;14(1):8-24. doi: 10.1631/jzus.B1200189. J Zhejiang Univ Sci B. 2013. PMID: 23303627 Free PMC article. Review.
-
EBV and Apoptosis: The Viral Master Regulator of Cell Fate?Viruses. 2017 Nov 13;9(11):339. doi: 10.3390/v9110339. Viruses. 2017. PMID: 29137176 Free PMC article. Review.
-
Inhibition of germinal centre apoptotic programmes by epstein-barr virus.Adv Hematol. 2011;2011:829525. doi: 10.1155/2011/829525. Epub 2011 Oct 23. Adv Hematol. 2011. PMID: 22110506 Free PMC article.
-
Burkitt's lymphoma: the Rosetta Stone deciphering Epstein-Barr virus biology.Semin Cancer Biol. 2009 Dec;19(6):377-88. doi: 10.1016/j.semcancer.2009.07.004. Epub 2009 Jul 18. Semin Cancer Biol. 2009. PMID: 19619657 Free PMC article. Review.
References
-
- Alfieri, C., M. Birkenbach, and E. Kieff. 1991. Early events in Epstein-Barr virus infection of human B lymphocytes. Virology 181:595-608. - PubMed
-
- Andersson, M. L., N. J. Stam, G. Klein, H. L. Ploegh, and M. G. Masucci. 1991. Aberrant expression of HLA class-I antigens in Burkitt lymphoma cells. Int. J. Cancer 47:544-550. - PubMed
-
- Arenzana-Seisdedos, F., P. Turpin, M. Rodriguez, D. Thomas, R. T. Hay, J. L. Virelizier, and C. Dargemont. 1997. Nuclear localization of I kappa B alpha promotes active transport of NF-kappa B from the nucleus to the cytoplasm. J. Cell Sci. 110:369-378. - PubMed
-
- Ben-Bassat, H., N. Goldblum, S. Mitrani, T. Goldblum, J. M. Yoffey, M. M. Cohen, Z. Bentwich, B. Ramot, E. Klein, and G. Klein. 1977. Establishment in continuous culture of a new type of lymphocyte from a “Burkitt like” malignant lymphoma (line D.G.-75). Int. J. Cancer 19:27-33. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

