Spatial organization of the mammalian genome surveillance machinery in response to DNA strand breaks
- PMID: 16618811
- PMCID: PMC2063811
- DOI: 10.1083/jcb.200510130
Spatial organization of the mammalian genome surveillance machinery in response to DNA strand breaks
Abstract
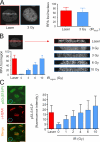
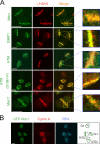
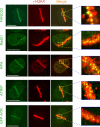
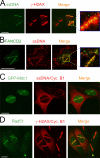
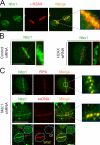
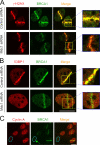
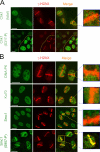
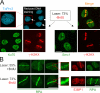
We show that DNA double-strand breaks (DSBs) induce complex subcompartmentalization of genome surveillance regulators. Chromatin marked by gamma-H2AX is occupied by ataxia telangiectasia-mutated (ATM) kinase, Mdc1, and 53BP1. In contrast, repair factors (Rad51, Rad52, BRCA2, and FANCD2), ATM and Rad-3-related (ATR) cascade (ATR, ATR interacting protein, and replication protein A), and the DNA clamp (Rad17 and -9) accumulate in subchromatin microcompartments delineated by single-stranded DNA (ssDNA). BRCA1 and the Mre11-Rad50-Nbs1 complex interact with both of these compartments. Importantly, some core DSB regulators do not form cytologically discernible foci. These are further subclassified to proteins that connect DSBs with the rest of the nucleus (Chk1 and -2), that assemble at unprocessed DSBs (DNA-PK/Ku70), and that exist on chromatin as preassembled complexes but become locally modified after DNA damage (Smc1/Smc3). Finally, checkpoint effectors such as p53 and Cdc25A do not accumulate at DSBs at all. We propose that subclassification of DSB regulators according to their residence sites provides a useful framework for understanding their involvement in diverse processes of genome surveillance.
Figures
Similar articles
-
53BP1 and NFBD1/MDC1-Nbs1 function in parallel interacting pathways activating ataxia-telangiectasia mutated (ATM) in response to DNA damage.Cancer Res. 2003 Dec 15;63(24):8586-91. Cancer Res. 2003. PMID: 14695167
-
ATM and the Mre11-Rad50-Nbs1 complex respond to nucleoside analogue-induced stalled replication forks and contribute to drug resistance.Cancer Res. 2008 Oct 1;68(19):7947-55. doi: 10.1158/0008-5472.CAN-08-0971. Cancer Res. 2008. PMID: 18829552 Free PMC article.
-
ATM regulates ATR chromatin loading in response to DNA double-strand breaks.J Exp Med. 2006 Feb 20;203(2):297-303. doi: 10.1084/jem.20051923. Epub 2006 Feb 6. J Exp Med. 2006. PMID: 16461339 Free PMC article.
-
The ATM-Chk2 and ATR-Chk1 pathways in DNA damage signaling and cancer.Adv Cancer Res. 2010;108:73-112. doi: 10.1016/B978-0-12-380888-2.00003-0. Adv Cancer Res. 2010. PMID: 21034966 Review.
-
Human syndromes with genomic instability and multiprotein machines that repair DNA double-strand breaks.Histol Histopathol. 2003 Jan;18(1):225-43. doi: 10.14670/HH-18.225. Histol Histopathol. 2003. PMID: 12507302 Review.
Cited by
-
Single-molecule imaging reveals the kinetics of non-homologous end-joining in living cells.bioRxiv [Preprint]. 2024 May 25:2023.06.22.546088. doi: 10.1101/2023.06.22.546088. bioRxiv. 2024. PMID: 38826211 Free PMC article. Preprint.
-
Decline of DNA damage response along with myogenic differentiation.Life Sci Alliance. 2023 Nov 22;7(2):e202302279. doi: 10.26508/lsa.202302279. Print 2024 Feb. Life Sci Alliance. 2023. PMID: 37993260 Free PMC article.
-
ATR promotes clearance of damaged DNA and damaged cells by rupturing micronuclei.Mol Cell. 2023 Oct 19;83(20):3642-3658.e4. doi: 10.1016/j.molcel.2023.09.003. Epub 2023 Oct 2. Mol Cell. 2023. PMID: 37788673 Free PMC article.
-
Live-cell tracking of γ-H2AX kinetics reveals the distinct modes of ATM and DNA-PK in the immediate response to DNA damage.J Cell Sci. 2023 Apr 15;136(8):jcs260698. doi: 10.1242/jcs.260698. Epub 2023 Apr 27. J Cell Sci. 2023. PMID: 36999484 Free PMC article.
-
TRIM24 is critical for the cellular response to DNA double-strand breaks through regulating the recruitment of MRN complex.Oncogene. 2023 Feb;42(8):586-600. doi: 10.1038/s41388-022-02580-8. Epub 2022 Dec 22. Oncogene. 2023. PMID: 36550358
References
-
- Abraham, R.T. 2004. PI 3-kinase related kinases: ‘big’ players in stress-induced signaling pathways. DNA Repair (Amst.). 3:883–887. - PubMed
-
- Aten, J.A., J. Stap, P.M. Krawczyk, C.H. van Oven, R.A. Hoebe, J. Essers, and R. Kanaar. 2004. Dynamics of DNA double-strand breaks revealed by clustering of damaged chromosome domains. Science. 303:92–95. - PubMed
-
- Bakkenist, C.J., and M.B. Kastan. 2003. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 421:499–506. - PubMed
-
- Celeste, A., O. Fernandez-Capetillo, M.J. Kruhlak, D.R. Pilch, D.W. Staudt, A. Lee, R.F. Bonner, W.M. Bonner, and A. Nussenzweig. 2003. Histone H2AX phosphorylation is dispensable for the initial recognition of DNA breaks. Nat. Cell Biol. 5:675–679. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

