Genome-wide relationships between TAF1 and histone acetyltransferases in Saccharomyces cerevisiae
- PMID: 16537921
- PMCID: PMC1430310
- DOI: 10.1128/MCB.26.7.2791-2802.2006
Genome-wide relationships between TAF1 and histone acetyltransferases in Saccharomyces cerevisiae
Abstract
Histone acetylation regulates gene expression, yet the functional contributions of the numerous histone acetyltransferases (HATs) to gene expression and their relationships with each other remain largely unexplored. The central role of the putative HAT-containing TAF1 subunit of TFIID in gene expression raises the fundamental question as to what extent, if any, TAF1 contributes to acetylation in vivo and to what extent it is redundant with other HATs. Our findings herein do not support the basic tenet that TAF1 is a major HAT in Saccharomyces cerevisiae, nor do we find that TAF1 is functionally redundant with other HATs, including Gcn5, Elp3, Hat1, Hpa2, Sas3, and Esa1, which is in contrast to previous conclusions regarding Gcn5. Our findings do reveal that of these HATs, only Gcn5 and Esa1 contribute substantially to gene expression genome wide. Interestingly, histone acetylation at promoter regions throughout the genome does not require TAF1 or RNA polymerase II, indicating that most acetylation is likely to precede transcription and not depend upon it. TAF1 function has been linked to Bdf1, which binds TFIID and acetylated histone H4 tails, but no linkage between TAF1 and the H4 HAT Esa1 has been established. Here, we present evidence for such a linkage through Bdf1.
Figures
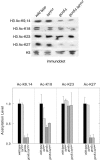
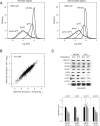
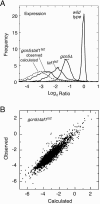

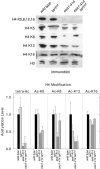

Similar articles
-
A role for gcn5-mediated global histone acetylation in transcriptional regulation.Mol Cell Biol. 2006 Mar;26(5):1610-6. doi: 10.1128/MCB.26.5.1610-1616.2006. Mol Cell Biol. 2006. PMID: 16478983 Free PMC article.
-
Differential requirement of SAGA subunits for Mot1p and Taf1p recruitment in gene activation.Mol Cell Biol. 2005 Jun;25(12):4863-72. doi: 10.1128/MCB.25.12.4863-4872.2005. Mol Cell Biol. 2005. PMID: 15923605 Free PMC article.
-
TAF1 histone acetyltransferase activity in Sp1 activation of the cyclin D1 promoter.Mol Cell Biol. 2005 May;25(10):4321-32. doi: 10.1128/MCB.25.10.4321-4332.2005. Mol Cell Biol. 2005. PMID: 15870300 Free PMC article.
-
Acetylation of histones and transcription-related factors.Microbiol Mol Biol Rev. 2000 Jun;64(2):435-59. doi: 10.1128/MMBR.64.2.435-459.2000. Microbiol Mol Biol Rev. 2000. PMID: 10839822 Free PMC article. Review.
-
Structure and chemistry of the p300/CBP and Rtt109 histone acetyltransferases: implications for histone acetyltransferase evolution and function.Curr Opin Struct Biol. 2008 Dec;18(6):741-7. doi: 10.1016/j.sbi.2008.09.004. Epub 2008 Oct 27. Curr Opin Struct Biol. 2008. PMID: 18845255 Free PMC article. Review.
Cited by
-
Functional dissection of the NuA4 histone acetyltransferase reveals its role as a genetic hub and that Eaf1 is essential for complex integrity.Mol Cell Biol. 2008 Apr;28(7):2244-56. doi: 10.1128/MCB.01653-07. Epub 2008 Jan 22. Mol Cell Biol. 2008. PMID: 18212056 Free PMC article.
-
A glycolytic burst drives glucose induction of global histone acetylation by picNuA4 and SAGA.Nucleic Acids Res. 2009 Jul;37(12):3969-80. doi: 10.1093/nar/gkp270. Epub 2009 Apr 30. Nucleic Acids Res. 2009. PMID: 19406923 Free PMC article.
-
Protein and DNA modifications: evolutionary imprints of bacterial biochemical diversification and geochemistry on the provenance of eukaryotic epigenetics.Cold Spring Harb Perspect Biol. 2014 Jul 1;6(7):a016063. doi: 10.1101/cshperspect.a016063. Cold Spring Harb Perspect Biol. 2014. PMID: 24984775 Free PMC article. Review.
-
A MYST family histone acetyltransferase, MoSAS3, is required for development and pathogenicity in the rice blast fungus.Mol Plant Pathol. 2019 Nov;20(11):1491-1505. doi: 10.1111/mpp.12856. Epub 2019 Jul 30. Mol Plant Pathol. 2019. PMID: 31364260 Free PMC article.
-
Features of cryptic promoters and their varied reliance on bromodomain-containing factors.PLoS One. 2010 Sep 23;5(9):e12927. doi: 10.1371/journal.pone.0012927. PLoS One. 2010. PMID: 20886085 Free PMC article.
References
-
- Ai, X., and M. R. Parthun. 2004. The nuclear Hat1p/Hat2p complex: a molecular link between type B histone acetyltransferases and chromatin assembly. Mol. Cell 14:195-205. - PubMed
-
- Basehoar, A. D., S. J. Zanton, and B. F. Pugh. 2004. Identification and distinct regulation of yeast TATA box-containing genes. Cell 116:699-709. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
