Thymic pathogenicity of an HIV-1 envelope is associated with increased CXCR4 binding efficiency and V5-gp41-dependent activity, but not V1/V2-associated CD4 binding efficiency and viral entry
- PMID: 15892960
- PMCID: PMC4415377
- DOI: 10.1016/j.virol.2005.03.032
Thymic pathogenicity of an HIV-1 envelope is associated with increased CXCR4 binding efficiency and V5-gp41-dependent activity, but not V1/V2-associated CD4 binding efficiency and viral entry
Abstract
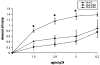
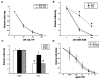
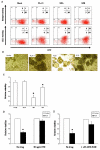
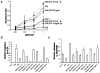
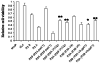
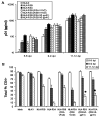
We previously described a thymus-tropic HIV-1 envelope (R3A Env) from a rapid progressor obtained at the time of transmission. An HIV-1 molecular recombinant with the R3A Env supported extensive replication and pathogenesis in the thymus and did not require Nef. Another Env from the same patient did not display the same thymus-tropic pathogenesis (R3B Env). Here, we show that relative to R3B Env, R3A Env enhances viral entry of T cells, increases fusion-induced cytopathicity, and shows elevated binding efficiency for both CD4 and CXCR4, but not CCR5, in vitro. We created chimeric envelopes to determine the region(s) responsible for each in vitro phenotype and for thymic pathogenesis. Surprisingly, while V1/V2 contributed to enhanced viral entry, CD4 binding efficiency, and cytopathicity in vitro, it made no contribution to thymic pathogenesis. Rather, CXCR4 binding efficiency and V5-gp41-associated activity appear to independently contribute to thymic pathogenesis of the R3A Env. These data highlight the contribution of unique HIV pathogenic factors in the thymic microenvironment and suggest that novel mechanisms may be involved in Env pathogenic activity in vivo.
Figures
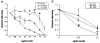

Similar articles
-
The heptad repeat 2 domain is a major determinant for enhanced human immunodeficiency virus type 1 (HIV-1) fusion and pathogenicity of a highly pathogenic HIV-1 Env.J Virol. 2009 Nov;83(22):11715-25. doi: 10.1128/JVI.00649-09. Epub 2009 Sep 2. J Virol. 2009. PMID: 19726524 Free PMC article.
-
Characterization of a thymus-tropic HIV-1 isolate from a rapid progressor: role of the envelope.Virology. 2004 Oct 10;328(1):74-88. doi: 10.1016/j.virol.2004.07.019. Virology. 2004. PMID: 15380360 Free PMC article.
-
CCR5 interaction with HIV-1 Env contributes to Env-induced depletion of CD4 T cells in vitro and in vivo.Retrovirology. 2016 Mar 29;13:22. doi: 10.1186/s12977-016-0255-z. Retrovirology. 2016. PMID: 27026376 Free PMC article.
-
Molecular Mechanism of HIV-1 Entry.Trends Microbiol. 2019 Oct;27(10):878-891. doi: 10.1016/j.tim.2019.06.002. Epub 2019 Jun 28. Trends Microbiol. 2019. PMID: 31262533 Free PMC article. Review.
-
Cellular entry of HIV: Evaluation of therapeutic targets.Curr Pharm Des. 2006;12(16):1963-73. doi: 10.2174/138161206777442155. Curr Pharm Des. 2006. PMID: 16787241 Review.
Cited by
-
HIV-1 infection and pathogenesis in a novel humanized mouse model.Blood. 2007 Apr 1;109(7):2978-81. doi: 10.1182/blood-2006-07-033159. Blood. 2007. PMID: 17132723 Free PMC article.
-
Derivation and characterization of a simian immunodeficiency virus SIVmac239 variant with tropism for CXCR4.J Virol. 2009 Oct;83(19):9911-22. doi: 10.1128/JVI.00533-09. Epub 2009 Jul 15. J Virol. 2009. PMID: 19605489 Free PMC article.
-
HIV-1 infection induces interleukin-1β production via TLR8 protein-dependent and NLRP3 inflammasome mechanisms in human monocytes.J Biol Chem. 2014 Aug 1;289(31):21716-26. doi: 10.1074/jbc.M114.566620. Epub 2014 Jun 17. J Biol Chem. 2014. PMID: 24939850 Free PMC article.
-
Identification of human microRNA-like sequences embedded within the protein-encoding genes of the human immunodeficiency virus.PLoS One. 2013;8(3):e58586. doi: 10.1371/journal.pone.0058586. Epub 2013 Mar 8. PLoS One. 2013. PMID: 23520522 Free PMC article.
-
Criteria for effective design, construction, and gene knockdown by shRNA vectors.BMC Biotechnol. 2006 Jan 24;6:7. doi: 10.1186/1472-6750-6-7. BMC Biotechnol. 2006. PMID: 16433925 Free PMC article.
References
-
- Arthos J, Cicala C, Selig SM, White AA, Ravindranath HM, Van Ryk D, Steenbeke TD, Machado E, Khazanie P, Hanback MS, Hanback DB, Rabin RL, Fauci AS. The role of the CD4 receptor versus HIV coreceptors in envelope-mediated apoptosis in peripheral blood mononuclear cells. Virology. 2002;292(1):98–106. - PubMed
-
- Beaumont T, Quakkelaar E, van Nuenen A, Pantophlet R, Schuitemaker H. Increased sensitivity to CD4 binding site-directed neutralization following in vitro propagation on primary lymphocytes of a neutralization-resistant human immunodeficiency virus IIIB strain isolated from an accidentally infected laboratory worker. J. Virol. 2004;78(11):5651–5657. - PMC - PubMed
-
- Berkowitz RD, Beckerman KP, Schall TJ, McCune JM. CXCR4 and CCR5 expression delineates targets for HIV-1 disruption of T cell differentiation. J. Immunol. 1998;161(7):3702–3710. - PubMed
-
- Bonyhadi ML, Su L, Auten J, McCune JM, Kaneshima H. Development of a human thymic organ culture model for the study of HIV pathogenesis. AIDS Res. Hum. Retroviruses. 1995;11(9):1073–1080. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

