Developmentally regulated alterations in Polycomb repressive complex 1 proteins on the inactive X chromosome
- PMID: 15596546
- PMCID: PMC2172612
- DOI: 10.1083/jcb.200409026
Developmentally regulated alterations in Polycomb repressive complex 1 proteins on the inactive X chromosome
Abstract
Polycomb group (PcG) proteins belonging to the polycomb (Pc) repressive complexes 1 and 2 (PRC1 and PRC2) maintain homeotic gene silencing. In Drosophila, PRC2 methylates histone H3 on lysine 27, and this epigenetic mark facilitates recruitment of PRC1. Mouse PRC2 (mPRC2) has been implicated in X inactivation, as mPRC2 proteins transiently accumulate on the inactive X chromosome (Xi) at the onset of X inactivation to methylate histone H3 lysine 27 (H3-K27). In this study, we demonstrate that mPRC1 proteins localize to the Xi, and that different mPRC1 proteins accumulate on the Xi during initiation and maintenance of X inactivation in embryonic cells. The Xi accumulation of mPRC1 proteins requires Xist RNA and is not solely regulated by the presence of H3-K27 methylation, as not all cells that exhibit this epigenetic mark on the Xi show Xi enrichment of mPRC1 proteins. Our results implicate mPRC1 in X inactivation and suggest that the regulated assembly of PcG protein complexes on the Xi contributes to this multistep process.
Figures
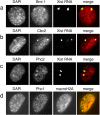
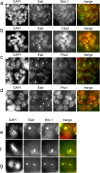
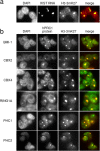
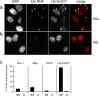

Similar articles
-
Role of histone H3 lysine 27 methylation in X inactivation.Science. 2003 Apr 4;300(5616):131-5. doi: 10.1126/science.1084274. Epub 2003 Mar 20. Science. 2003. PMID: 12649488
-
Role of histone H3 lysine 27 methylation in Polycomb-group silencing.Science. 2002 Nov 1;298(5595):1039-43. doi: 10.1126/science.1076997. Epub 2002 Sep 26. Science. 2002. PMID: 12351676
-
Stable X chromosome inactivation involves the PRC1 Polycomb complex and requires histone MACROH2A1 and the CULLIN3/SPOP ubiquitin E3 ligase.Proc Natl Acad Sci U S A. 2005 May 24;102(21):7635-40. doi: 10.1073/pnas.0408918102. Epub 2005 May 16. Proc Natl Acad Sci U S A. 2005. PMID: 15897469 Free PMC article.
-
The functions of E(Z)/EZH2-mediated methylation of lysine 27 in histone H3.Curr Opin Genet Dev. 2004 Apr;14(2):155-64. doi: 10.1016/j.gde.2004.02.001. Curr Opin Genet Dev. 2004. PMID: 15196462 Review.
-
Polycomb complexes in X chromosome inactivation.Philos Trans R Soc Lond B Biol Sci. 2017 Nov 5;372(1733):20170021. doi: 10.1098/rstb.2017.0021. Philos Trans R Soc Lond B Biol Sci. 2017. PMID: 28947664 Free PMC article. Review.
Cited by
-
Setdb1-mediated H3K9 methylation is enriched on the inactive X and plays a role in its epigenetic silencing.Epigenetics Chromatin. 2016 May 18;9:16. doi: 10.1186/s13072-016-0064-6. eCollection 2016. Epigenetics Chromatin. 2016. PMID: 27195021 Free PMC article.
-
Chromobox proteins in cancer: Multifaceted functions and strategies for modulation (Review).Int J Oncol. 2023 Mar;62(3):36. doi: 10.3892/ijo.2023.5484. Epub 2023 Feb 3. Int J Oncol. 2023. PMID: 36734270 Free PMC article. Review.
-
PRC1 collaborates with SMCHD1 to fold the X-chromosome and spread Xist RNA between chromosome compartments.Nat Commun. 2019 Jul 3;10(1):2950. doi: 10.1038/s41467-019-10755-3. Nat Commun. 2019. PMID: 31270318 Free PMC article.
-
Xist regulation and function explored.Hum Genet. 2011 Aug;130(2):223-36. doi: 10.1007/s00439-011-1008-7. Epub 2011 May 28. Hum Genet. 2011. PMID: 21626138 Free PMC article. Review.
-
A novel approach to differentiate rat embryonic stem cells in vitro reveals a role for RNF12 in activation of X chromosome inactivation.Sci Rep. 2019 Apr 15;9(1):6068. doi: 10.1038/s41598-019-42246-2. Sci Rep. 2019. PMID: 30988473 Free PMC article.
References
-
- Alkema, M.J., J. Jacobs, J.W. Voncken, N.A. Jenkins, N.G. Copeland, D.P. Satijn, A.P. Otte, A. Berns, and M. van Lohuizen. 1997. MPc2, a new murine homolog of the Drosophila polycomb protein is a member of the mouse polycomb transcriptional repressor complex. J. Mol. Biol. 273:993–1003. - PubMed
-
- Bardos, J.I., A.J. Saurin, C. Tissot, E. Duprez, and P.S. Freemont. 2000. HPC3 is a new human polycomb orthologue that interacts and associates with RING1 and Bmi1 and has transcriptional repression properties. J. Biol. Chem. 275:28785–28792. - PubMed
-
- Cao, R., and Y. Zhang. 2004. The functions of E(Z)/EZH2-mediated methylation of lysine 27 in histone H3. Curr. Opin. Genet. Dev. 14:155–164. - PubMed
-
- Cao, R., L. Wang, H. Wang, L. Xia, H. Erdjument-Bromage, P. Tempst, R.S. Jones, and Y. Zhang. 2002. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 298:1039–1043. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

