Recognition and cleavage of primary microRNA precursors by the nuclear processing enzyme Drosha
- PMID: 15565168
- PMCID: PMC544904
- DOI: 10.1038/sj.emboj.7600491
Recognition and cleavage of primary microRNA precursors by the nuclear processing enzyme Drosha
Abstract
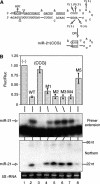
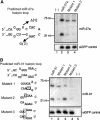
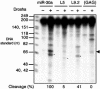
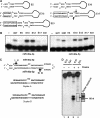
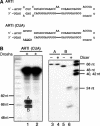
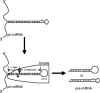
A critical step during human microRNA maturation is the processing of the primary microRNA transcript by the nuclear RNaseIII enzyme Drosha to generate the approximately 60-nucleotide precursor microRNA hairpin. How Drosha recognizes primary RNA substrates and selects its cleavage sites has remained a mystery, especially given that the known targets for Drosha processing show no discernable sequence homology. Here, we show that human Drosha selectively cleaves RNA hairpins bearing a large (>/=10 nucleotides) terminal loop. From the junction of the loop and the adjacent stem, Drosha then cleaves approximately two helical RNA turns into the stem to produce the precursor microRNA. Beyond the precursor microRNA cleavage sites, approximately one helix turn of stem extension is also essential for efficient processing. While the sites of Drosha cleavage are determined largely by the distance from the terminal loop, variations in stem structure and sequence around the cleavage site can fine-tune the actual cleavage sites chosen.
Figures



Similar articles
-
A central role for the primary microRNA stem in guiding the position and efficiency of Drosha processing of a viral pri-miRNA.RNA. 2014 Jul;20(7):1068-77. doi: 10.1261/rna.044537.114. Epub 2014 May 22. RNA. 2014. PMID: 24854622 Free PMC article.
-
Efficient processing of primary microRNA hairpins by Drosha requires flanking nonstructured RNA sequences.J Biol Chem. 2005 Jul 29;280(30):27595-603. doi: 10.1074/jbc.M504714200. Epub 2005 Jun 1. J Biol Chem. 2005. PMID: 15932881
-
Lower and upper stem-single-stranded RNA junctions together determine the Drosha cleavage site.Proc Natl Acad Sci U S A. 2013 Dec 17;110(51):20687-92. doi: 10.1073/pnas.1311639110. Epub 2013 Dec 2. Proc Natl Acad Sci U S A. 2013. PMID: 24297910 Free PMC article.
-
The role of the precursor structure in the biogenesis of microRNA.Cell Mol Life Sci. 2011 Sep;68(17):2859-71. doi: 10.1007/s00018-011-0726-2. Epub 2011 May 24. Cell Mol Life Sci. 2011. PMID: 21607569 Free PMC article. Review.
-
The double-stranded microRNA precursor.Postepy Biochem. 2024 May 23;70(1):57-61. doi: 10.18388/pb.2021_522. Print 2024 May 23. Postepy Biochem. 2024. PMID: 39016229 Review.
Cited by
-
miRNA arm selection and isomiR distribution in gastric cancer.BMC Genomics. 2012;13 Suppl 1(Suppl 1):S13. doi: 10.1186/1471-2164-13-S1-S13. Epub 2012 Jan 17. BMC Genomics. 2012. PMID: 22369582 Free PMC article.
-
Genetic variants in microRNA genes: impact on microRNA expression, function, and disease.Front Genet. 2015 May 21;6:186. doi: 10.3389/fgene.2015.00186. eCollection 2015. Front Genet. 2015. PMID: 26052338 Free PMC article. Review.
-
Computational identification of microRNA gene loci and precursor microRNA sequences in CHO cell lines.J Biotechnol. 2012 Apr 15;158(3):151-5. doi: 10.1016/j.jbiotec.2012.01.019. Epub 2012 Jan 25. J Biotechnol. 2012. PMID: 22306111 Free PMC article.
-
Altered dynamics of scaRNA2 and scaRNA9 in response to stress correlates with disrupted nuclear organization.Biol Open. 2018 Sep 27;7(9):bio037101. doi: 10.1242/bio.037101. Biol Open. 2018. PMID: 30177550 Free PMC article.
-
Correlation between sequence conservation and structural thermodynamics of microRNA precursors from human, mouse, and chicken genomes.BMC Evol Biol. 2010 Oct 27;10:329. doi: 10.1186/1471-2148-10-329. BMC Evol Biol. 2010. PMID: 20977776 Free PMC article.
References
-
- Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116: 281–297 - PubMed
-
- Brummelkamp TR, Bernards R, Agami R (2002) A system for stable expression of short interfering RNA in mammalian cells. Science 296: 550–553 - PubMed
-
- Carrington JC, Ambros V (2003) Role of microRNAs in plant and animal development. Science 301: 336–338 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

