Antagonistic interaction between abscisic acid and jasmonate-ethylene signaling pathways modulates defense gene expression and disease resistance in Arabidopsis
- PMID: 15548743
- PMCID: PMC535886
- DOI: 10.1105/tpc.104.025833
Antagonistic interaction between abscisic acid and jasmonate-ethylene signaling pathways modulates defense gene expression and disease resistance in Arabidopsis
Abstract
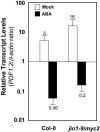
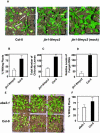
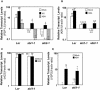
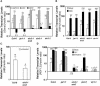
The plant hormones abscisic acid (ABA), jasmonic acid (JA), and ethylene are involved in diverse plant processes, including the regulation of gene expression during adaptive responses to abiotic and biotic stresses. Previously, ABA has been implicated in enhancing disease susceptibility in various plant species, but currently very little is known about the molecular mechanisms underlying this phenomenon. In this study, we obtained evidence that a complex interplay between ABA and JA-ethylene signaling pathways regulate plant defense gene expression and disease resistance. First, we showed that exogenous ABA suppressed both basal and JA-ethylene-activated transcription from defense genes. By contrast, ABA deficiency as conditioned by the mutations in the ABA1 and ABA2 genes, which encode enzymes involved in ABA biosynthesis, resulted in upregulation of basal and induced transcription from JA-ethylene responsive defense genes. Second, we found that disruption of AtMYC2 (allelic to JASMONATE INSENSITIVE1 [JIN1]), encoding a basic helix-loop-helix Leu zipper transcription factor, which is a positive regulator of ABA signaling, results in elevated levels of basal and activated transcription from JA-ethylene responsive defense genes. Furthermore, the jin1/myc2 and aba2-1 mutants showed increased resistance to the necrotrophic fungal pathogen Fusarium oxysporum. Finally, using ethylene and ABA signaling mutants, we showed that interaction between ABA and ethylene signaling is mutually antagonistic in vegetative tissues. Collectively, our results indicate that the antagonistic interactions between multiple components of ABA and the JA-ethylene signaling pathways modulate defense and stress responsive gene expression in response to biotic and abiotic stresses.
Figures


Similar articles
-
Heterotrimeric G proteins-mediated resistance to necrotrophic pathogens includes mechanisms independent of salicylic acid-, jasmonic acid/ethylene- and abscisic acid-mediated defense signaling.Plant J. 2009 Apr;58(1):69-81. doi: 10.1111/j.1365-313X.2008.03755.x. Epub 2008 Dec 29. Plant J. 2009. PMID: 19054360
-
Allantoin, a stress-related purine metabolite, can activate jasmonate signaling in a MYC2-regulated and abscisic acid-dependent manner.J Exp Bot. 2016 Apr;67(8):2519-2532. doi: 10.1093/jxb/erw071. Epub 2016 Mar 1. J Exp Bot. 2016. PMID: 26931169 Free PMC article.
-
JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis.Plant Cell. 2004 Jul;16(7):1938-50. doi: 10.1105/tpc.022319. Epub 2004 Jun 18. Plant Cell. 2004. PMID: 15208388 Free PMC article.
-
Signaling Crosstalk between Salicylic Acid and Ethylene/Jasmonate in Plant Defense: Do We Understand What They Are Whispering?Int J Mol Sci. 2019 Feb 4;20(3):671. doi: 10.3390/ijms20030671. Int J Mol Sci. 2019. PMID: 30720746 Free PMC article. Review.
-
Molecular basis for jasmonate and ethylene signal interactions in Arabidopsis.J Exp Bot. 2014 Nov;65(20):5743-8. doi: 10.1093/jxb/eru349. Epub 2014 Aug 27. J Exp Bot. 2014. PMID: 25165148 Review.
Cited by
-
Arabidopsis thaliana resistance to fusarium oxysporum 2 implicates tyrosine-sulfated peptide signaling in susceptibility and resistance to root infection.PLoS Genet. 2013 May;9(5):e1003525. doi: 10.1371/journal.pgen.1003525. Epub 2013 May 23. PLoS Genet. 2013. PMID: 23717215 Free PMC article.
-
Assessing HCH isomer uptake in Alnus glutinosa: implications for phytoremediation and microbial response.Sci Rep. 2024 Feb 20;14(1):4187. doi: 10.1038/s41598-024-54235-1. Sci Rep. 2024. PMID: 38378833 Free PMC article.
-
Genomics-assisted prediction of salt and alkali tolerances and functional marker development in apple rootstocks.BMC Genomics. 2020 Aug 10;21(1):550. doi: 10.1186/s12864-020-06961-9. BMC Genomics. 2020. PMID: 32778069 Free PMC article.
-
Characterization of Triticum aestivum Abscisic Acid Receptors and a Possible Role for These in Mediating Fusairum Head Blight Susceptibility in Wheat.PLoS One. 2016 Oct 18;11(10):e0164996. doi: 10.1371/journal.pone.0164996. eCollection 2016. PLoS One. 2016. PMID: 27755583 Free PMC article.
-
Central Metabolic Responses to Ozone and Herbivory Affect Photosynthesis and Stomatal Closure.Plant Physiol. 2016 Nov;172(3):2057-2078. doi: 10.1104/pp.16.01318. Epub 2016 Oct 6. Plant Physiol. 2016. PMID: 27758847 Free PMC article.
References
-
- Alonso, J.M., et al. (2003. a). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301, 653–657. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

