Amsacta moorei entomopoxvirus expresses an active superoxide dismutase
- PMID: 15367592
- PMCID: PMC516379
- DOI: 10.1128/JVI.78.19.10265-10275.2004
Amsacta moorei entomopoxvirus expresses an active superoxide dismutase
Abstract
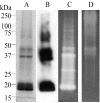
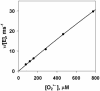
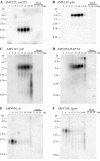
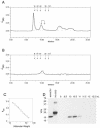
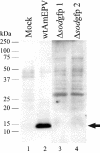
The entomopoxvirus from Amsacta moorei serves as the prototype of the group B entomopoxviruses. One of the interesting genes found in Amsacta moorei entomopoxvirus (AmEPV) is a superoxide dismutase (sod) (open reading frame AMV255). Superoxide dismutases (SODs) catalyze the conversion of superoxide radicals to hydrogen peroxide and oxygen. Many vertebrate poxviruses contain a sod gene, but to date, none have been demonstrated to be active. There are three families of SODs, characterized by their metal ion-binding partners, Fe, Mn, or Cu and Zn. Poxvirus enzymes belong to the Cu-Zn SOD family. Unlike inactive vertebrate poxvirus SODs, AMVSOD contains all the amino acids necessary for function. We expressed and purified a 6X-His-tagged version of the AMVSOD in Escherichia coli. The recombinant AMVSOD demonstrates superoxide dismutase activity both in an in situ gel assay and by stopped flow spectrophotometry. The k(cat)/K(m) for AMVSOD is 4 x 10(7) M(-1)s(-1). In infected cells, the AMVSOD protein behaves as a dimer and is catalytically active; however, disruption of the gene in AMEPV has little or no effect on growth of the virus in cell culture. An analysis of mRNA expression indicates that AMVsod is expressed late during infection of Lymantria dispar (Ld652) cells and produces a discrete nonpolydisperse transcript. Characterization of protein expression with a monoclonal antibody generated against AMVSOD confirms that the AMVSOD protein can be classified as a late, postreplicative gene. Therefore, AMVSOD is the first example of an active poxvirus SOD.
Figures


Similar articles
-
The effect of inhibitors on the growth of the entomopoxvirus from Amsacta moorei in Lymantria dispar (gypsy moth) cells.Virology. 1995 Aug 20;211(2):462-73. doi: 10.1006/viro.1995.1428. Virology. 1995. PMID: 7645250
-
Functional analysis of the inhibitor of apoptosis (iap) gene carried by the entomopoxvirus of Amsacta moorei.J Virol. 2005 Feb;79(4):2335-45. doi: 10.1128/JVI.79.4.2335-2345.2005. J Virol. 2005. PMID: 15681434 Free PMC article.
-
Complete genomic sequence of the Amsacta moorei entomopoxvirus: analysis and comparison with other poxviruses.Virology. 2000 Aug 15;274(1):120-39. doi: 10.1006/viro.2000.0449. Virology. 2000. PMID: 10936094
-
Characterization of two copper/zinc superoxide dismutases (Cu/Zn-SODs) from the desert beetle Microdera punctipennis and their activities in protecting E. coli cells against cold.Cryobiology. 2019 Apr;87:15-27. doi: 10.1016/j.cryobiol.2019.03.006. Epub 2019 Mar 16. Cryobiology. 2019. PMID: 30890324 Review.
-
Eukaryotic copper-only superoxide dismutases (SODs): A new class of SOD enzymes and SOD-like protein domains.J Biol Chem. 2018 Mar 30;293(13):4636-4643. doi: 10.1074/jbc.TM117.000182. Epub 2017 Dec 19. J Biol Chem. 2018. PMID: 29259135 Free PMC article. Review.
Cited by
-
Virologs, viral mimicry, and virocell metabolism: the expanding scale of cellular functions encoded in the complex genomes of giant viruses.FEMS Microbiol Rev. 2023 Sep 5;47(5):fuad053. doi: 10.1093/femsre/fuad053. FEMS Microbiol Rev. 2023. PMID: 37740576 Free PMC article.
-
Gypsy moth genome provides insights into flight capability and virus-host interactions.Proc Natl Acad Sci U S A. 2019 Jan 29;116(5):1669-1678. doi: 10.1073/pnas.1818283116. Epub 2019 Jan 14. Proc Natl Acad Sci U S A. 2019. PMID: 30642971 Free PMC article.
-
Host Range and Coding Potential of Eukaryotic Giant Viruses.Viruses. 2020 Nov 21;12(11):1337. doi: 10.3390/v12111337. Viruses. 2020. PMID: 33233432 Free PMC article. Review.
-
Preliminary crystallographic analysis of the Megavirus superoxide dismutase.Acta Crystallogr Sect F Struct Biol Cryst Commun. 2012 Dec 1;68(Pt 12):1557-9. doi: 10.1107/S174430911204657X. Epub 2012 Nov 28. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2012. PMID: 23192047 Free PMC article.
-
Chlorovirus PBCV-1 encodes an active copper-zinc superoxide dismutase.J Virol. 2014 Nov;88(21):12541-50. doi: 10.1128/JVI.02031-14. Epub 2014 Aug 20. J Virol. 2014. PMID: 25142578 Free PMC article.
References
-
- Abernethy, J. L., H. M. Steinman, and R. L. Hill. 1974. Bovine erythrocyte superoxide dismutase. Subunit structure and sequence location of the intrasubunit disulfide bond. J. Biol. Chem. 249:7339-7347. - PubMed
-
- Alscher, R. G., N. Erturk, and L. S. Heath. 2002. Role of superoxide dismutases (SODs) in controlling oxidative stress in plants. J. Exp. Bot. 53:1331-1341. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

