Accumulation of heterochromatin components on the terminal repeat sequence of Kaposi's sarcoma-associated herpesvirus mediated by the latency-associated nuclear antigen
- PMID: 15220403
- PMCID: PMC434099
- DOI: 10.1128/JVI.78.14.7299-7310.2004
Accumulation of heterochromatin components on the terminal repeat sequence of Kaposi's sarcoma-associated herpesvirus mediated by the latency-associated nuclear antigen
Abstract
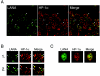
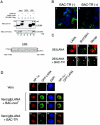
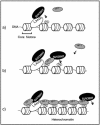
In the latent infection of Kaposi's sarcoma-associated herpesvirus (KSHV), its 160-kb circularized episomal DNA is replicated and maintained in the host nucleus. KSHV latency-associated nuclear antigen (LANA) is a key factor for maintaining viral latency. LANA binds to the terminal repeat (TR) DNA of the viral genome, leading to its localization to specific dot structures in the nucleus. In such an infected cell, the expression of the viral genes is restricted by a mechanism that is still unclear. Here, we found that LANA interacts with SUV39H1 histone methyltransferase, a key component of heterochromatin formation, as determined by use of a DNA pull-down assay with a biotinylated DNA fragment that contained a LANA-specific binding sequence and a maltose-binding protein pull-down assay. The diffuse localization of LANA on the chromosomes of uninfected cells changed to a punctate one with the introduction of a bacterial artificial chromosome containing most of the TR region, and SUV39H1 clearly colocalized with the LANA-associated dots. Thus, the LANA foci in KSHV-infected cells seemed to include SUV39H1 as well as heterochromatin protein 1. Furthermore, a chromatin immunoprecipitation assay revealed that the TR and the open reading frame (ORF) K1 and ORF50/RTA genes, but not the ORF73/LANA gene, lay within the heterochromatin during KSHV latency. Taken together, these observations indicate that LANA recruits heterochromatin components to the viral genome, which may lead to the establishment of viral latency and govern the transcription program.
Figures



Similar articles
-
Kaposi's sarcoma-associated herpesvirus-encoded latency-associated nuclear antigen modulates K1 expression through its cis-acting elements within the terminal repeats.J Virol. 2006 Apr;80(7):3445-58. doi: 10.1128/JVI.80.7.3445-3458.2006. J Virol. 2006. PMID: 16537612 Free PMC article.
-
Promoter switching allows simultaneous transcription of LANA and K14/vGPCR of Kaposi's sarcoma-associated herpesvirus.Virology. 2006 Jun 20;350(1):192-205. doi: 10.1016/j.virol.2006.03.006. Epub 2006 Apr 17. Virology. 2006. PMID: 16616289
-
Accumulation of LANA at nuclear matrix fraction is important for Kaposi's sarcoma-associated herpesvirus replication in latency.Virus Res. 2009 Jan;139(1):74-84. doi: 10.1016/j.virusres.2008.10.011. Epub 2008 Dec 9. Virus Res. 2009. PMID: 19027806
-
Lytic cycle switches of oncogenic human gammaherpesviruses.Adv Cancer Res. 2007;97:81-109. doi: 10.1016/S0065-230X(06)97004-3. Adv Cancer Res. 2007. PMID: 17419942 Review.
-
KSHV LANA--the master regulator of KSHV latency.Viruses. 2014 Dec 11;6(12):4961-98. doi: 10.3390/v6124961. Viruses. 2014. PMID: 25514370 Free PMC article. Review.
Cited by
-
Kruppel-associated box domain-associated protein-1 as a latency regulator for Kaposi's sarcoma-associated herpesvirus and its modulation by the viral protein kinase.Cancer Res. 2009 Jul 15;69(14):5681-9. doi: 10.1158/0008-5472.CAN-08-4570. Epub 2009 Jul 7. Cancer Res. 2009. PMID: 19584288 Free PMC article.
-
The latency-associated nuclear antigen interacts with MeCP2 and nucleosomes through separate domains.J Virol. 2010 Mar;84(5):2318-30. doi: 10.1128/JVI.01097-09. Epub 2009 Dec 23. J Virol. 2010. PMID: 20032179 Free PMC article.
-
Viral latency and its regulation: lessons from the gamma-herpesviruses.Cell Host Microbe. 2010 Jul 22;8(1):100-15. doi: 10.1016/j.chom.2010.06.014. Cell Host Microbe. 2010. PMID: 20638646 Free PMC article. Review.
-
KSHV LANA inhibits TGF-beta signaling through epigenetic silencing of the TGF-beta type II receptor.Blood. 2008 May 1;111(9):4731-40. doi: 10.1182/blood-2007-09-110544. Epub 2008 Jan 16. Blood. 2008. PMID: 18199825 Free PMC article.
-
Comprehensive analysis of LANA interacting proteins essential for viral genome tethering and persistence.PLoS One. 2013 Sep 11;8(9):e74662. doi: 10.1371/journal.pone.0074662. eCollection 2013. PLoS One. 2013. PMID: 24040311 Free PMC article.
References
-
- Aagaard, L., G. Laible, P. Selenko, M. Schmid, R. Dorn, G. Schotta, S. Kuhfittig, A. Wolf, A. Lebersorger, P. B. Singh, G. Reuter, and T. Jenuwein. 1999. Functional mammalian homologues of the Drosophila PEV-modifier Su(var)3-9 encode centromere-associated proteins which complex with the heterochromatin component M31. EMBO J. 18:1923-1938. - PMC - PubMed
-
- Ballestas, M. E., P. A. Chatis, and K. M. Kaye. 1999. Efficient persistence of extrachromosomal KSHV DNA mediated by latency-associated nuclear antigen. Science 284:641-644. - PubMed
-
- Bannister, A. J., P. Zegerman, J. F. Partridge, E. A. Miska, J. O. Thomas, R. C. Allshire, and T. Kouzarides. 2001. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410:120-124. - PubMed
-
- Bell, A. C., A. G. West, and G. Felsenfeld. 1999. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell 98:387-396. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Research Materials

