Identification of Krüppel-like factor 4 as a potential tumor suppressor gene in colorectal cancer
- PMID: 14724568
- PMCID: PMC1351029
- DOI: 10.1038/sj.onc.1207067
Identification of Krüppel-like factor 4 as a potential tumor suppressor gene in colorectal cancer
Abstract
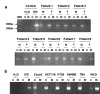
Krüppel-like factor 4 (KLF4 or GKLF) is an inhibitor of the cell cycle. The gene encoding KLF4 is localized on chromosome 9q, previously shown to exhibit allelic loss in colorectal cancer (CRC). In this study, we show that the mean level of KLF4 mRNA in a panel of 30 CRC was 52% that of paired normal colonic tissues. Similarly, the levels of KLF4 mRNA and protein in a panel of six established CRC cell lines were significantly lower than those of an untransformed colonic epithelial cell line. Using highly polymorphic DNA markers that flank the KLF4 locus, we found evidence for loss of heterozygosity (LOH) in two of eight surgically resected CRC specimens. In addition, LOH was observed in five of six CRC cell lines with one additional cell line exhibiting hemizygous deletion in the KLF4 gene. We also found that the 5'-untranslated region of KLF4 was hypermethylated in a subset of resected CRC specimens and cell lines. Lastly, the open-reading frame of KLF4 in two of three CRC cell lines examined contained several point mutations that resulted in a diminished ability to activate the p21(WAF1/Cip1) promoter. These findings indicate that KLF4 is a potential tumor suppressor gene in CRC.
Figures
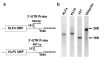





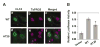
Similar articles
-
STAT1 is required for IFN-gamma-mediated gut-enriched Krüppel-like factor expression.Exp Cell Res. 2002 Nov 15;281(1):19-27. doi: 10.1006/excr.2002.5633. Exp Cell Res. 2002. PMID: 12441126
-
Downregulation and growth inhibitory effect of epithelial-type Krüppel-like transcription factor KLF4, but not KLF5, in bladder cancer.Biochem Biophys Res Commun. 2003 Aug 22;308(2):251-6. doi: 10.1016/s0006-291x(03)01356-1. Biochem Biophys Res Commun. 2003. PMID: 12901861
-
The gut-enriched Kruppel-like factor (Kruppel-like factor 4) mediates the transactivating effect of p53 on the p21WAF1/Cip1 promoter.J Biol Chem. 2000 Jun 16;275(24):18391-8. doi: 10.1074/jbc.C000062200. J Biol Chem. 2000. PMID: 10749849 Free PMC article.
-
Involvement of the multiple tumor suppressor genes and 12-lipoxygenase in human prostate cancer. Therapeutic implications.Adv Exp Med Biol. 1997;407:41-53. doi: 10.1007/978-1-4899-1813-0_7. Adv Exp Med Biol. 1997. PMID: 9321930 Review.
-
KLF4, p21 and context-dependent opposing forces in cancer.Nat Rev Cancer. 2006 Jan;6(1):11-23. doi: 10.1038/nrc1780. Nat Rev Cancer. 2006. PMID: 16372018 Review.
Cited by
-
[Correlation of KLF4 and SPARC expression with the clinical characteristics of non-small cell lung cancer].Zhongguo Fei Ai Za Zhi. 2012 Dec;15(12):720-4. doi: 10.3779/j.issn.1009-3419.2012.12.05. Zhongguo Fei Ai Za Zhi. 2012. PMID: 23249717 Free PMC article. Chinese.
-
Deoxycholic Acid Upregulates the Reprogramming Factors KFL4 and OCT4 Through the IL-6/STAT3 Pathway in Esophageal Adenocarcinoma Cells.Technol Cancer Res Treat. 2020 Jan-Dec;19:1533033820945302. doi: 10.1177/1533033820945302. Technol Cancer Res Treat. 2020. PMID: 32869704 Free PMC article.
-
Nuclear factor I-C regulates E-cadherin via control of KLF4 in breast cancer.BMC Cancer. 2015 Mar 10;15:113. doi: 10.1186/s12885-015-1118-z. BMC Cancer. 2015. PMID: 25879941 Free PMC article.
-
Phase 1 study of APTO-253 HCl, an inducer of KLF4, in patients with advanced or metastatic solid tumors.Invest New Drugs. 2015 Oct;33(5):1086-92. doi: 10.1007/s10637-015-0273-z. Epub 2015 Aug 14. Invest New Drugs. 2015. PMID: 26268924 Clinical Trial.
-
Induced pluripotent stem cell-related genes influence biological behavior and 5-fluorouracil sensitivity of colorectal cancer cells.J Zhejiang Univ Sci B. 2012 Jan;13(1):11-9. doi: 10.1631/jzus.B1100154. J Zhejiang Univ Sci B. 2012. PMID: 22205615 Free PMC article.
References
-
- American Cancer Society, Cancer Facts and Figures. 2003. (http://www.cancer.org/downloads/STT/CAFF2003PWSecured.pdf).
-
- Baker SJ, Markowitz S, Fearon ER, Willson JK, Vogelstein B. Science. 1990a;249:912–915. - PubMed
-
- Baker SJ, Preisinger AC, Jessup JM, Paraskeva C, Markowitz S, Willson JK, Hamilton S, Vogelstein B. Cancer Res. 1990b;50:7717–7722. - PubMed
-
- Bieker JJ. J Biol Chem. 2001;276:34355–34358. - PubMed
-
- Bird AP. Cancer Surv. 1996;28:87–101. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

