Genetic and functional analysis of full-length human immunodeficiency virus type 1 env genes derived from brain and blood of patients with AIDS
- PMID: 14581570
- PMCID: PMC254258
- DOI: 10.1128/jvi.77.22.12336-12345.2003
Genetic and functional analysis of full-length human immunodeficiency virus type 1 env genes derived from brain and blood of patients with AIDS
Abstract
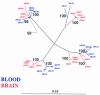
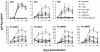
The genetic evolution of human immunodeficiency virus type 1 (HIV-1) in the brain is distinct from that in lymphoid tissues, indicating tissue-specific compartmentalization of the virus. Few primary HIV-1 envelope glycoproteins (Envs) from uncultured brain tissues have been biologically well characterized. In this study, we analyzed 37 full-length env genes from uncultured brain biopsy and blood samples from four patients with AIDS. Phylogenetic analysis of intrapatient sequence sets showed distinct clustering of brain relative to blood env sequences. However, no brain-specific signature sequence was identified. Furthermore, there was no significant difference in the number or positions of N-linked glycosylation sites between brain and blood env sequences. The patterns of coreceptor usage were heterogeneous, with no clear distinction between brain and blood env clones. Nine Envs used CCR5 as a coreceptor, one used CXCR4, and two used both CCR5 and CXCR4 in cell-to-cell fusion assays. Eight Envs could also use CCR3, CCR8, GPR15, STRL33, Apj, and/or GPR1, but these coreceptors did not play a major role in virus entry into microglia. Recognition of epitopes by the 2F5, T30, AG10H9, F105, 17b, and C11 monoclonal antibodies varied among env clones, reflecting genetic and conformational heterogeneity. Envs from two patients contained 28 to 32 N-glycosylation sites in gp120, compared to around 25 in lab strains and well-characterized primary isolates. These results suggest that HIV-1 Envs in brain cannot be distinguished from those in blood on the basis of coreceptor usage or the number or positions of N-glycosylation sites, indicating that other properties underlie neurotropism. The study also demonstrates characteristics of primary HIV-1 Envs from uncultured tissues and implies that Env variants that are glycosylated more extensively than lab strains and well-characterized primary isolates should be considered during development of vaccines and neutralizing antibodies.
Figures


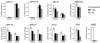
Similar articles
-
Patterns of chemokine receptor fusion cofactor utilization by human immunodeficiency virus type 1 variants from the lungs and blood.J Virol. 1999 Aug;73(8):6680-90. doi: 10.1128/JVI.73.8.6680-6690.1999. J Virol. 1999. PMID: 10400765 Free PMC article.
-
Macrophage tropism of human immunodeficiency virus type 1 isolates from brain and lymphoid tissues predicts neurotropism independent of coreceptor specificity.J Virol. 2001 Nov;75(21):10073-89. doi: 10.1128/JVI.75.21.10073-10089.2001. J Virol. 2001. PMID: 11581376 Free PMC article.
-
Tissue-specific sequence alterations in the human immunodeficiency virus type 1 envelope favoring CCR5 usage contribute to persistence of dual-tropic virus in the brain.J Virol. 2009 Jun;83(11):5430-41. doi: 10.1128/JVI.02648-08. Epub 2009 Mar 25. J Virol. 2009. PMID: 19321618 Free PMC article.
-
Genotypic coreceptor analysis.Eur J Med Res. 2007 Oct 15;12(9):453-62. Eur J Med Res. 2007. PMID: 17933727 Review.
-
Centralized HIV-1 envelope immunogens and neutralizing antibodies.Curr HIV Res. 2007 Nov;5(6):572-7. doi: 10.2174/157016207782418498. Curr HIV Res. 2007. PMID: 18045113 Review.
Cited by
-
The Glutamate System as a Crucial Regulator of CNS Toxicity and Survival of HIV Reservoirs.Front Cell Infect Microbiol. 2020 Jun 24;10:261. doi: 10.3389/fcimb.2020.00261. eCollection 2020. Front Cell Infect Microbiol. 2020. PMID: 32670889 Free PMC article. Review.
-
A machine learning approach for identifying amino acid signatures in the HIV env gene predictive of dementia.PLoS One. 2012;7(11):e49538. doi: 10.1371/journal.pone.0049538. Epub 2012 Nov 14. PLoS One. 2012. PMID: 23166702 Free PMC article.
-
Compartmentalized replication of R5 T cell-tropic HIV-1 in the central nervous system early in the course of infection.PLoS Pathog. 2015 Mar 26;11(3):e1004720. doi: 10.1371/journal.ppat.1004720. eCollection 2015 Mar. PLoS Pathog. 2015. PMID: 25811757 Free PMC article. Clinical Trial.
-
Phylodynamic analysis of human immunodeficiency virus type 1 in distinct brain compartments provides a model for the neuropathogenesis of AIDS.J Virol. 2005 Sep;79(17):11343-52. doi: 10.1128/JVI.79.17.11343-11352.2005. J Virol. 2005. PMID: 16103186 Free PMC article.
-
Simian immunodeficiency virus envelope compartmentalizes in brain regions independent of neuropathology.J Neurovirol. 2006 Apr;12(2):73-89. doi: 10.1080/13550280600654565. J Neurovirol. 2006. PMID: 16798669
References
-
- Albright, A. V., J. T. C. Shieh, T. Itoh, B. Lee, D. Pleasure, M. J. O'Connor, R. W. Doms, and F. Gonzalez-Scarano. 1999. Microglia express CCR5, CXCR4, and CCR3, but of these, CCR5 is the principal coreceptor for human immunodeficiency virus type 1 dementia isolates. J. Virol. 73:205-213. - PMC - PubMed
-
- Berger, E. A., P. M. Murphy, and J. M. Farber. 1999. Chemokine receptors as HIV-1 coreceptors: roles in viral entry, tropism, and disease. Annu. Rev. Immunol. 17:657-700. - PubMed
-
- Buchacher, A., R. Predl, K. Strutzenberger, W. Steinfellner, A. Trkola, M. Purtscher, G. Gruber, C. Tauer, F. Steindl, A. Jungbauer, and H. Katinger. 1994. Generation of human monoclonal antibodies against HIV-1 proteins; electrofusion and Epstein-Barr virus transformation for peripheral blood lymphocyte immortalization. AIDS Res. Hum. Retrovir. 10:359-369. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

