The double bromodomain protein Brd4 binds to acetylated chromatin during interphase and mitosis
- PMID: 12840145
- PMCID: PMC166386
- DOI: 10.1073/pnas.1433065100
The double bromodomain protein Brd4 binds to acetylated chromatin during interphase and mitosis
Abstract
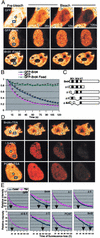
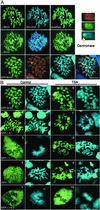
Previous in vitro studies showed that the bromodomain binds to acetyllysines on histone tails, leading to the proposal that the domain is involved in deciphering the histone code. However, there is little in vivo evidence supporting the binding of bromodomains to acetylated chromatin in the native environment. Brd4 is a member of the BET family that carries two bromodomains. It associates with mitotic chromosomes, a feature characteristic of the family. Here, we studied the interaction of Brd4 with chromatin in living cells by photobleaching. Brd4 was mobile and interacted with chromatin with a rapid "on and off" mode of binding. This interaction required both bromodomains. Indicating a preferential interaction with acetylated chromatin, Brd4 became less mobile upon increased chromatin acetylation caused by a histone deacetylase inhibitor. Providing biochemical support, salt solubility of Brd4 was markedly reduced upon increased histone acetylation. This change also required both bromodomains. In peptide binding assays, Brd4 avidly bound to di- and tetraacetylated histone H4 and diacetylated H3, but weakly or not at all to mono- and unacetylated H3 and H4. By contrast, it did not bind to unacetylated H4 or H3. Further, Brd4 colocalized with acetylated H4 and H3 in noncentromeric regions of mitotic chromosomes. This colocalization also required both bromodomains. These observations indicate that Brd4 specifically recognizes acetylated histone codes, and this recognition is passed onto the chromatin of newly divided cells.
Figures


Similar articles
-
Structural basis and binding properties of the second bromodomain of Brd4 with acetylated histone tails.Biochemistry. 2008 Jun 17;47(24):6403-17. doi: 10.1021/bi8001659. Epub 2008 May 24. Biochemistry. 2008. PMID: 18500820
-
Selective recognition of acetylated histones by bromodomains in transcriptional co-activators.Biochem J. 2007 Feb 15;402(1):125-33. doi: 10.1042/BJ20060907. Biochem J. 2007. PMID: 17049045 Free PMC article.
-
Interaction of bovine papillomavirus E2 protein with Brd4 stabilizes its association with chromatin.J Virol. 2005 Jul;79(14):8920-32. doi: 10.1128/JVI.79.14.8920-8932.2005. J Virol. 2005. PMID: 15994786 Free PMC article.
-
Do protein motifs read the histone code?Bioessays. 2005 Feb;27(2):164-75. doi: 10.1002/bies.20176. Bioessays. 2005. PMID: 15666348 Review.
-
Biological function and histone recognition of family IV bromodomain-containing proteins.J Cell Physiol. 2018 Mar;233(3):1877-1886. doi: 10.1002/jcp.26010. Epub 2017 Jun 13. J Cell Physiol. 2018. PMID: 28500727 Free PMC article. Review.
Cited by
-
Bromodomain-Containing Protein BRD4 Is Hyperphosphorylated in Mitosis.Cancers (Basel). 2020 Jun 20;12(6):1637. doi: 10.3390/cancers12061637. Cancers (Basel). 2020. PMID: 32575711 Free PMC article.
-
The BET family member BRD4 interacts with OCT4 and regulates pluripotency gene expression.Stem Cell Reports. 2015 Mar 10;4(3):390-403. doi: 10.1016/j.stemcr.2015.01.012. Epub 2015 Feb 12. Stem Cell Reports. 2015. PMID: 25684227 Free PMC article.
-
PFI-1, a highly selective protein interaction inhibitor, targeting BET Bromodomains.Cancer Res. 2013 Jun 1;73(11):3336-46. doi: 10.1158/0008-5472.CAN-12-3292. Epub 2013 Apr 10. Cancer Res. 2013. PMID: 23576556 Free PMC article.
-
High-Resolution Mapping of RNA Polymerases Identifies Mechanisms of Sensitivity and Resistance to BET Inhibitors in t(8;21) AML.Cell Rep. 2016 Aug 16;16(7):2003-16. doi: 10.1016/j.celrep.2016.07.032. Epub 2016 Aug 4. Cell Rep. 2016. PMID: 27498870 Free PMC article.
-
Stress-induced nuclear depletion of Entamoeba histolytica 3'-5' exoribonuclease EhRrp6 and its role in growth and erythrophagocytosis.J Biol Chem. 2018 Oct 19;293(42):16242-16260. doi: 10.1074/jbc.RA118.004632. Epub 2018 Aug 31. J Biol Chem. 2018. PMID: 30171071 Free PMC article.
References
-
- Strahl, B. D. & Allis, C. D. (2000) Nature 403, 41–45. - PubMed
-
- Jenuwein, T. & Allis, C. D. (2001) Science 293, 1074–1080. - PubMed
-
- Jeanmougin, F., Wurtz, J. M., Le Douarin, B., Chambon, P. & Losson, R. (1997) Trends Biochem. Sci. 22, 151–153. - PubMed
-
- Dhalluin, C., Carlson, J. E., Zeng, L., He, C., Aggarwal, A. K. & Zhou, M. M. (1999) Nature 399, 491–496. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

