Complement regulation by Kaposi's sarcoma-associated herpesvirus ORF4 protein
- PMID: 12477863
- PMCID: PMC140610
- DOI: 10.1128/jvi.77.1.592-599.2003
Complement regulation by Kaposi's sarcoma-associated herpesvirus ORF4 protein
Abstract
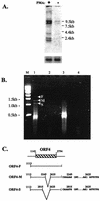
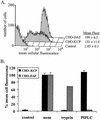
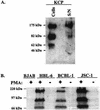
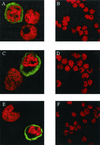
Kaposi's sarcoma-associated herpesvirus (KSHV) is associated with three types of human tumor: Kaposi's sarcoma, multicentric Castleman's disease, and primary effusion lymphoma. The virus encodes a number of proteins that participate in disrupting the immune response, one of which was predicted by sequence analysis to be encoded by open reading frame 4 (ORF4). The predicted ORF4 protein shares homology with cellular proteins referred to as regulators of complement activation. In the present study, the transcription profile of the ORF4 gene was characterized, revealing that it encodes at least three transcripts, by alternative splicing mechanisms, and three protein isoforms. Functional studies revealed that each ORF4 protein isoform inhibits complement and retains a C-terminal transmembrane domain. Consistent with the complement-regulating activity, we propose to name the proteins encoded by the ORF4 gene collectively as KSHV complement control protein (KCP). KSHV ORF4 is the most complex alternatively spliced gene encoding a viral complement regulator described to date. KCP inhibits the complement component of the innate immune response, thereby possibly contributing to the in vivo persistence and pathogenesis of this virus.
Figures



Similar articles
-
Functional activity of the complement regulator encoded by Kaposi's sarcoma-associated herpesvirus.J Biol Chem. 2003 Mar 14;278(11):9283-9. doi: 10.1074/jbc.m211579200. J Biol Chem. 2003. PMID: 12645526
-
Identification of a spliced gene from Kaposi's sarcoma-associated herpesvirus encoding a protein with similarities to latent membrane proteins 1 and 2A of Epstein-Barr virus.J Virol. 1999 Aug;73(8):6953-63. doi: 10.1128/JVI.73.8.6953-6963.1999. J Virol. 1999. PMID: 10400794 Free PMC article.
-
Molecular characterization of the rhesus rhadinovirus (RRV) ORF4 gene and the RRV complement control protein it encodes.J Virol. 2007 Apr;81(8):4166-76. doi: 10.1128/JVI.02069-06. Epub 2007 Feb 7. J Virol. 2007. PMID: 17287274 Free PMC article.
-
Modulation of cell signaling pathways by Kaposi's sarcoma-associated herpesvirus (KSHVHHV-8).Cell Biochem Biophys. 2004;40(3):305-22. doi: 10.1385/CBB:40:3:305. Cell Biochem Biophys. 2004. PMID: 15211030 Review.
-
Pathological Features of Kaposi's Sarcoma-Associated Herpesvirus Infection.Adv Exp Med Biol. 2018;1045:357-376. doi: 10.1007/978-981-10-7230-7_16. Adv Exp Med Biol. 2018. PMID: 29896675 Review.
Cited by
-
Probing the interaction between feline immunodeficiency virus and CD134 by using the novel monoclonal antibody 7D6 and the CD134 (Ox40) ligand.J Virol. 2007 Sep;81(18):9665-79. doi: 10.1128/JVI.01020-07. Epub 2007 Jul 3. J Virol. 2007. PMID: 17609274 Free PMC article.
-
The relevance of complement to virus biology.Virology. 2004 Feb 20;319(2):176-84. doi: 10.1016/j.virol.2003.11.029. Virology. 2004. PMID: 15015499 Free PMC article. Review. No abstract available.
-
Dissecting the regions of virion-associated Kaposi's sarcoma-associated herpesvirus complement control protein required for complement regulation and cell binding.J Virol. 2006 Apr;80(8):4068-78. doi: 10.1128/JVI.80.8.4068-4078.2006. J Virol. 2006. PMID: 16571823 Free PMC article.
-
Glycosaminoglycan interactions in murine gammaherpesvirus-68 infection.PLoS One. 2007 Apr 4;2(4):e347. doi: 10.1371/journal.pone.0000347. PLoS One. 2007. PMID: 17406671 Free PMC article.
-
Enforced covalent trimerisation of soluble feline CD134 (OX40)-ligand generates a functional antagonist of feline immunodeficiency virus.Mol Immunol. 2009 Mar;46(6):1020-30. doi: 10.1016/j.molimm.2008.08.271. Epub 2009 Feb 1. Mol Immunol. 2009. PMID: 19181384 Free PMC article.
References
-
- Albrecht, J. C., J. Nicholas, K. R. Cameron, C. Newman, B. Fleckenstein, and R. W. Honess. 1992. Herpesvirus saimiri has a gene specifying a homologue of the cellular membrane glycoprotein CD59. Virology 190:527-530. - PubMed
-
- Alexander, L., L. Denekamp, A. Knapp, M. R. Auerbach, B. Damania, and R. C. Desrosiers. 2000. The primary sequence of rhesus monkey rhadinovirus isolate 26-95: sequence similarities to Kaposi's sarcoma-associated herpesvirus and rhesus monkey rhadinovirus isolate 17577. J. Virol. 74:3388-3398. - PMC - PubMed
-
- Blackbourn, D. J., L. F. Chuang, S. Sutjipto, K. F. Killam, Jr., P. M. McCready, R. H. Doi, Y. Li, and R. Y. Chuang. 1992. Detection of simian immunodeficiency virus RNA from infected rhesus macaques by the polymerase chain reaction. J. Virol. Methods 37:109-117. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

