Sensitivity of HIV-1 to entry inhibitors correlates with envelope/coreceptor affinity, receptor density, and fusion kinetics
- PMID: 12444251
- PMCID: PMC138597
- DOI: 10.1073/pnas.252469399
Sensitivity of HIV-1 to entry inhibitors correlates with envelope/coreceptor affinity, receptor density, and fusion kinetics
Abstract
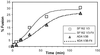
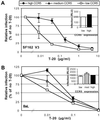
HIV entry inhibitors include coreceptor antagonists and the fusion inhibitor T-20. T-20 binds the first helical region (HR1) in the gp41 subunit of the viral envelope (Env) protein and prevents conformational changes required for membrane fusion. HR1 appears to become accessible to T-20 after Env binds CD4, whereas coreceptor binding is thought to induce the final conformational changes that lead to membrane fusion. Thus, T-20 binds to a structural intermediate of the fusion process. Primary viruses exhibit considerable variability in T-20 sensitivity, and determinants outside of HR1 can affect sensitivity by unknown mechanisms. We studied chimeric Env proteins containing different V3 loop sequences and found that gp120coreceptor affinity correlated with T-20 and coreceptor antagonist sensitivity, with greater affinity resulting in increased resistance to both classes of entry inhibitors. Enhanced affinity resulted in more rapid fusion kinetics, reducing the time during which Env is sensitive to T-20. Reduced coreceptor expression levels also delayed fusion kinetics and enhanced virus sensitivity to T-20, whereas increased coreceptor levels had the opposite effect. A single amino acid change (K421D) in the bridging sheet region of the primary virus strain YU2 reduced affinity for CCR5 and increased T-20 sensitivity by about 30-fold. Thus, mutations in Env that affect receptor engagement and membrane fusion rates can alter entry inhibitor sensitivity. Because coreceptor expression levels are typically limiting in vivo, individuals who express lower coreceptor levels may respond more favorably to entry inhibitors such as T-20, whose effectiveness we show depends in part on fusion kinetics.
Figures
Similar articles
-
Kinetic factors control efficiencies of cell entry, efficacies of entry inhibitors, and mechanisms of adaptation of human immunodeficiency virus.J Virol. 2005 Apr;79(7):4347-56. doi: 10.1128/JVI.79.7.4347-4356.2005. J Virol. 2005. PMID: 15767435 Free PMC article.
-
Sensitivity of human immunodeficiency virus type 1 to fusion inhibitors targeted to the gp41 first heptad repeat involves distinct regions of gp41 and is consistently modulated by gp120 interactions with the coreceptor.J Virol. 2001 Sep;75(18):8605-14. doi: 10.1128/jvi.75.18.8605-8614.2001. J Virol. 2001. PMID: 11507206 Free PMC article.
-
Impact of mutations in the coreceptor binding site on human immunodeficiency virus type 1 fusion, infection, and entry inhibitor sensitivity.J Virol. 2004 May;78(10):5476-85. doi: 10.1128/jvi.78.10.5476-5485.2004. J Virol. 2004. PMID: 15113926 Free PMC article.
-
HIV entry inhibitors: mechanisms of action and resistance pathways.J Antimicrob Chemother. 2006 Apr;57(4):619-27. doi: 10.1093/jac/dkl027. Epub 2006 Feb 7. J Antimicrob Chemother. 2006. PMID: 16464888 Review.
-
Progress in targeting HIV-1 entry.Drug Discov Today. 2005 Aug 15;10(16):1085-94. doi: 10.1016/S1359-6446(05)03550-6. Drug Discov Today. 2005. PMID: 16182193 Review.
Cited by
-
HIV-1 diversity in the envelope glycoproteins: implications for viral entry inhibition.Viruses. 2013 Feb 6;5(2):595-604. doi: 10.3390/v5020595. Viruses. 2013. PMID: 23389465 Free PMC article. Review.
-
Emerging drug targets for antiretroviral therapy.Drugs. 2005;65(13):1747-66. doi: 10.2165/00003495-200565130-00002. Drugs. 2005. PMID: 16114975 Review.
-
Human immunodeficiency virus (HIV) gp41 escape mutants: cross-resistance to peptide inhibitors of HIV fusion and altered receptor activation of gp120.J Virol. 2005 Apr;79(8):4774-81. doi: 10.1128/JVI.79.8.4774-4781.2005. J Virol. 2005. PMID: 15795263 Free PMC article.
-
An antibody directed against the fusion peptide of Junin virus envelope glycoprotein GPC inhibits pH-induced membrane fusion.J Virol. 2010 Jun;84(12):6119-29. doi: 10.1128/JVI.02700-09. Epub 2010 Apr 14. J Virol. 2010. PMID: 20392854 Free PMC article.
-
The appealing story of HIV entry inhibitors : from discovery of biological mechanisms to drug development.Drugs. 2005;65(7):879-904. doi: 10.2165/00003495-200565070-00001. Drugs. 2005. PMID: 15892586
References
-
- Trkola A., Dragic, T., Arthos, J., Binley, J. M., Olson, W. C., Allaway, G. P., Cheng-Meyer, C., Robinson, J., Maddon, P. J. & Moore, J. P. (1996) Nature 384, 184-187. - PubMed
-
- Wu L., Gerard, N. P., Wyatt, R., Choe, H., Parolin, C., Ruffing, N., Borsetti, A., Cardoso, A. A., Desjardin, E., Newman, W., et al. (1996) Nature 384, 179-183. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

