Cellular transcriptional profiling in influenza A virus-infected lung epithelial cells: the role of the nonstructural NS1 protein in the evasion of the host innate defense and its potential contribution to pandemic influenza
- PMID: 12149435
- PMCID: PMC125029
- DOI: 10.1073/pnas.112338099
Cellular transcriptional profiling in influenza A virus-infected lung epithelial cells: the role of the nonstructural NS1 protein in the evasion of the host innate defense and its potential contribution to pandemic influenza
Abstract
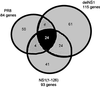
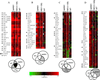
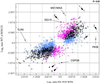
The NS1 protein of influenza A virus contributes to viral pathogenesis, primarily by enabling the virus to disarm the host cell type IFN defense system. We examined the downstream effects of NS1 protein expression during influenza A virus infection on global cellular mRNA levels by measuring expression of over 13,000 cellular genes in response to infection with wild-type and mutant viruses in human lung epithelial cells. Influenza A/PR/8/34 virus infection resulted in a significant induction of genes involved in the IFN pathway. Deletion of the viral NS1 gene increased the number and magnitude of expression of cellular genes implicated in the IFN, NF-kappaB, and other antiviral pathways. Interestingly, different IFN-induced genes showed different sensitivities to NS1-mediated inhibition of their expression. A recombinant virus with a C-terminal deletion in its NS1 gene induced an intermediate cellular mRNA expression pattern between wild-type and NS1 knockout viruses. Most significantly, a virus containing the 1918 pandemic NS1 gene was more efficient at blocking the expression of IFN-regulated genes than its parental influenza A/WSN/33 virus. Taken together, our results suggest that the cellular response to influenza A virus infection in human lung cells is significantly influenced by the sequence of the NS1 gene, demonstrating the importance of the NS1 protein in regulating the host cell response triggered by virus infection.
Figures
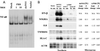

Similar articles
-
Influenza A virus protein PB1-F2 exacerbates IFN-beta expression of human respiratory epithelial cells.J Immunol. 2010 Oct 15;185(8):4812-23. doi: 10.4049/jimmunol.0903952. Epub 2010 Sep 15. J Immunol. 2010. PMID: 20844191
-
Differential Modulation of Innate Immune Responses in Human Primary Cells by Influenza A Viruses Carrying Human or Avian Nonstructural Protein 1.J Virol. 2019 Dec 12;94(1):e00999-19. doi: 10.1128/JVI.00999-19. Print 2019 Dec 12. J Virol. 2019. PMID: 31597767 Free PMC article.
-
Influenza A virus NS1 protein prevents activation of NF-kappaB and induction of alpha/beta interferon.J Virol. 2000 Dec;74(24):11566-73. doi: 10.1128/jvi.74.24.11566-11573.2000. J Virol. 2000. PMID: 11090154 Free PMC article.
-
Modulation of Innate Immune Responses by the Influenza A NS1 and PA-X Proteins.Viruses. 2018 Dec 12;10(12):708. doi: 10.3390/v10120708. Viruses. 2018. PMID: 30545063 Free PMC article. Review.
-
The influenza virus NS1 protein: inhibitor of innate and adaptive immunity.Infect Disord Drug Targets. 2007 Dec;7(4):336-43. doi: 10.2174/187152607783018754. Infect Disord Drug Targets. 2007. PMID: 18220965 Review.
Cited by
-
Transcriptomic comparison of primary human lung cells with lung tissue samples and the human A549 lung cell line highlights cell type specific responses during infections with influenza A virus.Sci Rep. 2022 Nov 29;12(1):20608. doi: 10.1038/s41598-022-24792-4. Sci Rep. 2022. PMID: 36446841 Free PMC article.
-
18S rRNA is a reliable normalisation gene for real time PCR based on influenza virus infected cells.Virol J. 2012 Oct 8;9:230. doi: 10.1186/1743-422X-9-230. Virol J. 2012. PMID: 23043930 Free PMC article.
-
The Nucleocapsid Protein of Coronaviruses Acts as a Viral Suppressor of RNA Silencing in Mammalian Cells.J Virol. 2015 Sep;89(17):9029-43. doi: 10.1128/JVI.01331-15. Epub 2015 Jun 17. J Virol. 2015. PMID: 26085159 Free PMC article.
-
Influenza infection directly alters innate IL-23 and IL-12p70 and subsequent IL-17A and IFN-γ responses to pneumococcus in vitro in human monocytes.PLoS One. 2018 Sep 7;13(9):e0203521. doi: 10.1371/journal.pone.0203521. eCollection 2018. PLoS One. 2018. PMID: 30192848 Free PMC article.
-
Pathogenicity of influenza viruses with genes from the 1918 pandemic virus: functional roles of alveolar macrophages and neutrophils in limiting virus replication and mortality in mice.J Virol. 2005 Dec;79(23):14933-44. doi: 10.1128/JVI.79.23.14933-14944.2005. J Virol. 2005. PMID: 16282492 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

