Terminal nucleotidyl transferase activity of recombinant Flaviviridae RNA-dependent RNA polymerases: implication for viral RNA synthesis
- PMID: 11507207
- PMCID: PMC115107
- DOI: 10.1128/jvi.75.18.8615-8623.2001
Terminal nucleotidyl transferase activity of recombinant Flaviviridae RNA-dependent RNA polymerases: implication for viral RNA synthesis
Abstract
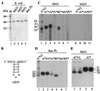
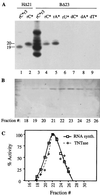
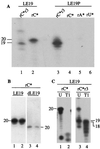
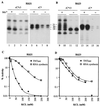
Recombinant hepatitis C virus (HCV) RNA-dependent RNA polymerase (RdRp) was reported to possess terminal transferase (TNTase) activity, the ability to add nontemplated nucleotides to the 3' end of viral RNAs. However, this TNTase was later purported to be a cellular enzyme copurifying with the HCV RdRp. In this report, we present evidence that TNTase activity is an inherent function of HCV and bovine viral diarrhea virus RdRps highly purified from both prokaryotic and eukaryotic cells. A change of the highly conserved GDD catalytic motif in the HCV RdRp to GAA abolished both RNA synthesis and TNTase activity. Furthermore, the nucleotides added via this TNTase activity are strongly influenced by the sequence near the 3' terminus of the viral template RNA, perhaps accounting for the previous discrepant observations between RdRp preparations. Last, the RdRp TNTase activity was shown to restore the ability to direct initiation of RNA synthesis in vitro on an initiation-defective RNA substrate, thereby implicating this activity in maintaining the integrity of the viral genome termini.
Figures
Similar articles
-
The RNA-dependent RNA polymerases of different members of the family Flaviviridae exhibit similar properties in vitro.J Gen Virol. 1999 Oct;80 ( Pt 10):2583-2590. doi: 10.1099/0022-1317-80-10-2583. J Gen Virol. 1999. PMID: 10573150
-
Evidence for Internal Initiation of RNA Synthesis by the Hepatitis C Virus RNA-Dependent RNA Polymerase NS5B In Cellulo.J Virol. 2019 Sep 12;93(19):e00525-19. doi: 10.1128/JVI.00525-19. Print 2019 Oct 1. J Virol. 2019. PMID: 31315989 Free PMC article.
-
A recombinant hepatitis C virus RNA-dependent RNA polymerase capable of copying the full-length viral RNA.J Virol. 1999 Sep;73(9):7694-702. doi: 10.1128/JVI.73.9.7694-7702.1999. J Virol. 1999. PMID: 10438859 Free PMC article.
-
Hepatitis C virus RNA-dependent RNA polymerase (NS5B polymerase).Curr Top Microbiol Immunol. 2000;242:225-60. doi: 10.1007/978-3-642-59605-6_11. Curr Top Microbiol Immunol. 2000. PMID: 10592663 Review. No abstract available.
-
Biochemical and structural analysis of the NS5B RNA-dependent RNA polymerase of the hepatitis C virus.J Viral Hepat. 2000 May;7(3):167-74. doi: 10.1046/j.1365-2893.2000.00218.x. J Viral Hepat. 2000. PMID: 10849258 Review.
Cited by
-
De novo initiation pocket mutations have multiple effects on hepatitis C virus RNA-dependent RNA polymerase activities.J Virol. 2004 Nov;78(22):12207-17. doi: 10.1128/JVI.78.22.12207-12217.2004. J Virol. 2004. PMID: 15507607 Free PMC article.
-
De novo synthesis of negative-strand RNA by Dengue virus RNA-dependent RNA polymerase in vitro: nucleotide, primer, and template parameters.J Virol. 2003 Aug;77(16):8831-42. doi: 10.1128/jvi.77.16.8831-8842.2003. J Virol. 2003. PMID: 12885902 Free PMC article.
-
Ebolavirus polymerase uses an unconventional genome replication mechanism.Proc Natl Acad Sci U S A. 2019 Apr 23;116(17):8535-8543. doi: 10.1073/pnas.1815745116. Epub 2019 Apr 8. Proc Natl Acad Sci U S A. 2019. PMID: 30962389 Free PMC article.
-
Identification and Characterization of a Human Coronavirus 229E Nonstructural Protein 8-Associated RNA 3'-Terminal Adenylyltransferase Activity.J Virol. 2019 May 29;93(12):e00291-19. doi: 10.1128/JVI.00291-19. Print 2019 Jun 15. J Virol. 2019. PMID: 30918070 Free PMC article.
-
Arenavirus Z protein controls viral RNA synthesis by locking a polymerase-promoter complex.Proc Natl Acad Sci U S A. 2011 Dec 6;108(49):19743-8. doi: 10.1073/pnas.1112742108. Epub 2011 Nov 21. Proc Natl Acad Sci U S A. 2011. PMID: 22106304 Free PMC article.
References
-
- Al R H, Xie Y, Wang Y, Hagedorn C H. Expression of recombinant hepatitis C virus non-structural protein 5B in Escherichia coli. Virus Res. 1998;53:141–149. - PubMed
-
- Arnold J J, Ghosh S K B, Cameron C E. Poliovirus RNA-dependent RNA polymerase (3Dpol) J Biol Chem. 1999;274:37060–37069. - PubMed
-
- Beckman M T, Kirkegaard K. Site size of cooperative single-stranded RNA binding by poliovirus RNA-dependent RNA polymerase. J Biol Chem. 1998;273:6724–6730. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

