Human TLR9 confers responsiveness to bacterial DNA via species-specific CpG motif recognition
- PMID: 11470918
- PMCID: PMC55404
- DOI: 10.1073/pnas.161293498
Human TLR9 confers responsiveness to bacterial DNA via species-specific CpG motif recognition
Abstract
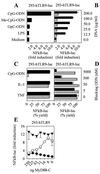
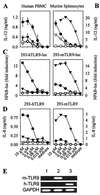
The Toll-like receptor (TLR) family consists of phylogenetically conserved transmembrane proteins, which function as mediators of innate immunity for recognition of pathogen-derived ligands and subsequent cell activation via the Toll/IL-1R signal pathway. Here, we show that human TLR9 (hTLR9) expression in human immune cells correlates with responsiveness to bacterial deoxycytidylate-phosphate-deoxyguanylate (CpG)-DNA. Notably "gain of function" to immunostimulatory CpG-DNA is achieved by expressing TLR9 in human nonresponder cells. Transfection of either human or murine TLR9 conferred responsiveness in a CD14- and MD2-independent manner, yet required species-specific CpG-DNA motifs for initiation of the Toll/IL-1R signal pathway via MyD88. The optimal CpG motif for hTLR9 was GTCGTT, whereas the optimal murine sequence was GACGTT. Overall, these data suggest that hTLR9 conveys CpG-DNA responsiveness to human cells by directly engaging immunostimulating CpG-DNA.
Figures


Similar articles
-
A Toll-like receptor recognizes bacterial DNA.Nature. 2000 Dec 7;408(6813):740-5. doi: 10.1038/35047123. Nature. 2000. PMID: 11130078
-
Toll-like receptor 9 mediates CpG-DNA signaling.J Leukoc Biol. 2002 Mar;71(3):538-44. J Leukoc Biol. 2002. PMID: 11867692
-
TLR9 signals after translocating from the ER to CpG DNA in the lysosome.Nat Immunol. 2004 Feb;5(2):190-8. doi: 10.1038/ni1028. Epub 2004 Jan 11. Nat Immunol. 2004. PMID: 14716310
-
Bovine toll-like receptor 9: a comparative analysis of molecular structure, function and expression.Vet Immunol Immunopathol. 2005 Oct 18;108(1-2):11-6. doi: 10.1016/j.vetimm.2005.07.012. Vet Immunol Immunopathol. 2005. PMID: 16098606 Review.
-
Recent advances in the development of immunostimulatory oligonucleotides.Curr Opin Drug Discov Devel. 2003 Mar;6(2):204-17. Curr Opin Drug Discov Devel. 2003. PMID: 12669456 Review.
Cited by
-
CpG Adjuvant in Allergen-Specific Immunotherapy: Finding the Sweet Spot for the Induction of Immune Tolerance.Front Immunol. 2021 Feb 23;12:590054. doi: 10.3389/fimmu.2021.590054. eCollection 2021. Front Immunol. 2021. PMID: 33708195 Free PMC article. Review.
-
The Trinity of cGAS, TLR9, and ALRs Guardians of the Cellular Galaxy Against Host-Derived Self-DNA.Front Immunol. 2021 Feb 11;11:624597. doi: 10.3389/fimmu.2020.624597. eCollection 2020. Front Immunol. 2021. PMID: 33643304 Free PMC article. Review.
-
Cancer immunotherapy employing an innovative strategy to enhance CD4+ T cell help in the tumor microenvironment.PLoS One. 2014 Dec 22;9(12):e115711. doi: 10.1371/journal.pone.0115711. eCollection 2014. PLoS One. 2014. PMID: 25531529 Free PMC article.
-
Adjuvant Effect of Toll-Like Receptor 9 Activation on Cancer Immunotherapy Using Checkpoint Blockade.Front Immunol. 2020 May 29;11:1075. doi: 10.3389/fimmu.2020.01075. eCollection 2020. Front Immunol. 2020. PMID: 32547560 Free PMC article. Review.
-
T-bet+ B cells are activated by and control endogenous retroviruses through TLR-dependent mechanisms.Nat Commun. 2024 Feb 9;15(1):1229. doi: 10.1038/s41467-024-45201-6. Nat Commun. 2024. PMID: 38336876 Free PMC article.
References
-
- Medzhitov R, Janeway C A., Jr Curr Opin Immunol. 1997;9:4–9. - PubMed
-
- Aderem A, Ulevitch R J. Nature (London) 2000;406:782–787. - PubMed
-
- Medzhitov R, Preston-Hurlburt P, Kopp E, Stadlen A, Chen C, Ghosh S, Janeway C A., Jr Mol Cell. 1998;2:253–258. - PubMed
-
- Muzio M, Polentarutti N, Bosisio D, Manoj Kumar P P, Mantovani A. Biochem Soc Trans. 2000;28:563–566. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

