Establishment of an oriP replicon is dependent upon an infrequent, epigenetic event
- PMID: 11390644
- PMCID: PMC87076
- DOI: 10.1128/MCB.21.13.4149-4161.2001
Establishment of an oriP replicon is dependent upon an infrequent, epigenetic event
Abstract
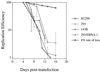
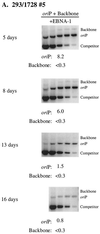
Plasmids containing oriP, the latent origin of replication for Epstein-Barr virus, support efficient replication in selected cell clones when the viral protein EBNA-1 is provided, being lost at a rate of 2 to 4% per cell generation after removal of selection (A. L. Kirchmaier and B. Sugden, J. Virol. 69:1280-1283, 1995; B. Sugden and N. Warren, Mol. Biol. Med. 5:85-94, 1988). We refer to these plasmids as established replicons in that they support efficient DNA synthesis and partitioning each cell cycle. Unexpectedly, we have found that upon introduction of oriP plasmids into a population of EBNA-1-positive cells, oriP plasmids replicate but are lost precipitously from cells during 2 weeks posttransfection (>25% rate of loss per cell generation). Upon investigation of these disparate observations, we have found that only 1 to 10% of cells transfected with an oriP plasmid expressing EBNA-1 and hygromycin phosphotransferase give rise to drug-resistant clones in which the oriP replicon is established. A hereditable alteration in these drug-resistant cell clones, manifested at the genetic or epigenetic level, does not underlie the establishment of oriP, as newly introduced oriP plasmids replicate but are also lost rapidly from these cells. In addition, a genetic alteration in the oriP plasmid is not responsible for establishment, as oriP plasmids isolated from an established cell clone, propagated in Escherichia coli, and reintroduced into EBNA-1-positive cells are likewise established inefficiently. Our findings demonstrate that oriP replicons are not intrinsically stable in EBNA-1-positive cell lines. Rather, the establishment of an oriP replicon is conferred upon the replicon by a stochastic, epigenetic event that occurs infrequently and, therefore, is detected in only a minority of cells.
Figures









Similar articles
-
The plasmid replicon of Epstein-Barr virus: mechanistic insights into efficient, licensed, extrachromosomal replication in human cells.Plasmid. 2007 Jul;58(1):1-12. doi: 10.1016/j.plasmid.2007.01.003. Epub 2007 Mar 9. Plasmid. 2007. PMID: 17350094 Free PMC article. Review.
-
The cis-acting family of repeats can inhibit as well as stimulate establishment of an oriP replicon.J Virol. 2001 Nov;75(22):10709-20. doi: 10.1128/JVI.75.22.10709-10720.2001. J Virol. 2001. PMID: 11602712 Free PMC article.
-
The plasmid replicon of EBV consists of multiple cis-acting elements that facilitate DNA synthesis by the cell and a viral maintenance element.EMBO J. 1998 Nov 2;17(21):6394-403. doi: 10.1093/emboj/17.21.6394. EMBO J. 1998. PMID: 9799247 Free PMC article.
-
The minimal replicator of Epstein-Barr virus oriP.J Virol. 2000 May;74(10):4512-22. doi: 10.1128/jvi.74.10.4512-4522.2000. J Virol. 2000. PMID: 10775587 Free PMC article.
-
Replication licensing of the EBV oriP minichromosome.Curr Top Microbiol Immunol. 2001;258:13-33. doi: 10.1007/978-3-642-56515-1_2. Curr Top Microbiol Immunol. 2001. PMID: 11443858 Review.
Cited by
-
pEPito: a significantly improved non-viral episomal expression vector for mammalian cells.BMC Biotechnol. 2010 Mar 15;10:20. doi: 10.1186/1472-6750-10-20. BMC Biotechnol. 2010. PMID: 20230618 Free PMC article.
-
Identification of new human origins of DNA replication by an origin-trapping assay.Mol Cell Biol. 2006 Oct;26(20):7731-46. doi: 10.1128/MCB.01392-06. Epub 2006 Sep 5. Mol Cell Biol. 2006. PMID: 16954389 Free PMC article.
-
Identification of properties of the Kaposi's sarcoma-associated herpesvirus latent origin of replication that are essential for the efficient establishment and maintenance of intact plasmids.J Virol. 2014 Aug;88(15):8490-503. doi: 10.1128/JVI.00742-14. Epub 2014 May 14. J Virol. 2014. PMID: 24829342 Free PMC article.
-
Nuclear import of Epstein-Barr virus nuclear antigen 1 mediated by NPI-1 (Importin alpha5) is up- and down-regulated by phosphorylation of the nuclear localization signal for which Lys379 and Arg380 are essential.J Virol. 2006 Feb;80(4):1979-91. doi: 10.1128/JVI.80.4.1979-1991.2006. J Virol. 2006. PMID: 16439554 Free PMC article.
-
The plasmid replicon of Epstein-Barr virus: mechanistic insights into efficient, licensed, extrachromosomal replication in human cells.Plasmid. 2007 Jul;58(1):1-12. doi: 10.1016/j.plasmid.2007.01.003. Epub 2007 Mar 9. Plasmid. 2007. PMID: 17350094 Free PMC article. Review.
References
-
- Aiyar A, Sugden B. Fusions between Epstein-Barr viral nuclear antigen-1 of Epstein-Barr virus and the large T-antigen of simian virus 40 replicate their cognate origins. J Biol Chem. 1998;273:33073–33081. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
