An unexpectedly high excision capacity for mispaired 5-hydroxymethyluracil in human cell extracts
- PMID: 11121024
- PMCID: PMC18892
- DOI: 10.1073/pnas.97.26.14183
An unexpectedly high excision capacity for mispaired 5-hydroxymethyluracil in human cell extracts
Abstract
The oxidation of thymine in DNA can generate a base pair between 5-hydroxymethyluracil (HmU) and adenine, whereas the oxidation and deamination of 5-methylcytosine (5mC) in DNA can generate a base pair between HmU and guanine. Using synthetic oligonucleotides containing HmU at a defined site, HmU-DNA glycosylase activities in HeLa cell and human fibroblast cell extracts have been observed. An HmU-DNA glycosylase activity that removes HmU mispaired with guanine has been measured. Surprisingly, the HmU:G excision activity is 60 times greater than the corresponding HmU:A activity, even though the expected rate of formation of the HmU:A base pair exceeds that of the HmU:G base pair by a factor of 10(7). The HmU:G mispair would arise from the 5mC:G base pair, and, if unrepaired, would give rise to a transition mutation. The observation of an unexpectedly high HmU:G glycosylase activity suggests that human cells may encounter the HmU:G mispair much more frequently than expected. The conversion of 5mC to HmU must be considered as a potential pathway for the generation of 5mC to T transition mutations, which are often found in human tumors.
Figures
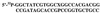
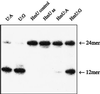
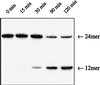

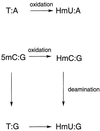

Similar articles
-
Identification of high excision capacity for 5-hydroxymethyluracil mispaired with guanine in DNA of Escherichia coli MutM, Nei and Nth DNA glycosylases.Nucleic Acids Res. 2003 Feb 15;31(4):1191-6. doi: 10.1093/nar/gkg223. Nucleic Acids Res. 2003. PMID: 12582238 Free PMC article.
-
Characterization of the substrate specificity of a human 5-hydroxymethyluracil glycosylase activity.Chem Res Toxicol. 2002 Jan;15(1):33-9. doi: 10.1021/tx010113b. Chem Res Toxicol. 2002. PMID: 11800595
-
Reduced 5-hydroxymethyluracil-DNA glycosylase activity in Werner's syndrome cells.Mutat Res. 1992 Mar;275(2):87-96. doi: 10.1016/0921-8734(92)90012-e. Mutat Res. 1992. PMID: 1379342
-
Enigmatic 5-hydroxymethyluracil: Oxidatively modified base, epigenetic mark or both?Mutat Res Rev Mutat Res. 2016 Jan-Mar;767:59-66. doi: 10.1016/j.mrrev.2016.02.001. Epub 2016 Feb 9. Mutat Res Rev Mutat Res. 2016. PMID: 27036066 Review.
-
Repair of deaminated bases in DNA.Free Radic Biol Med. 2002 Oct 1;33(7):886-93. doi: 10.1016/s0891-5849(02)00902-4. Free Radic Biol Med. 2002. PMID: 12361800 Review.
Cited by
-
DNA glycosylases: in DNA repair and beyond.Chromosoma. 2012 Feb;121(1):1-20. doi: 10.1007/s00412-011-0347-4. Epub 2011 Nov 3. Chromosoma. 2012. PMID: 22048164 Free PMC article. Review.
-
An interplay of the base excision repair and mismatch repair pathways in active DNA demethylation.Nucleic Acids Res. 2016 May 5;44(8):3713-27. doi: 10.1093/nar/gkw059. Epub 2016 Feb 3. Nucleic Acids Res. 2016. PMID: 26843430 Free PMC article.
-
Germline ablation of SMUG1 DNA glycosylase causes loss of 5-hydroxymethyluracil- and UNG-backup uracil-excision activities and increases cancer predisposition of Ung-/-Msh2-/- mice.Nucleic Acids Res. 2012 Jul;40(13):6016-25. doi: 10.1093/nar/gks259. Epub 2012 Mar 24. Nucleic Acids Res. 2012. PMID: 22447450 Free PMC article.
-
Detection of Cytosine methylation in ancient DNA from five native american populations using bisulfite sequencing.PLoS One. 2015 May 27;10(5):e0125344. doi: 10.1371/journal.pone.0125344. eCollection 2015. PLoS One. 2015. PMID: 26016479 Free PMC article.
-
Occurrence, Biological Consequences, and Human Health Relevance of Oxidative Stress-Induced DNA Damage.Chem Res Toxicol. 2016 Dec 19;29(12):2008-2039. doi: 10.1021/acs.chemrestox.6b00265. Epub 2016 Nov 7. Chem Res Toxicol. 2016. PMID: 27989142 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

