The protein encoded by the proto-oncogene DEK changes the topology of chromatin and reduces the efficiency of DNA replication in a chromatin-specific manner
- PMID: 10837023
- PMCID: PMC316669
The protein encoded by the proto-oncogene DEK changes the topology of chromatin and reduces the efficiency of DNA replication in a chromatin-specific manner
Abstract
The structure of chromatin regulates the genetic activity of the underlying DNA sequence. We report here that the protein encoded by the proto-oncogene DEK, which is involved in acute myelogenous leukemia, induces alterations of the superhelical density of DNA in chromatin. The change in topology is observed with chromatin but not with naked DNA and does not involve dissociation of core histones from chromatin. Moreover, these effects require histone H2A/H2B dimers in addition to histone H3/H4. We additionally tested whether the DEK protein affects DNA-utilizing processes and found that the DEK protein substantially reduces the replication efficiency of chromatin but not of naked DNA templates.
Figures

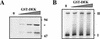

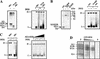
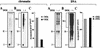
Similar articles
-
Daxx and histone deacetylase II associate with chromatin through an interaction with core histones and the chromatin-associated protein Dek.J Cell Sci. 2002 Aug 15;115(Pt 16):3319-30. doi: 10.1242/jcs.115.16.3319. J Cell Sci. 2002. PMID: 12140263
-
Bacterial Growth Inhibition Screen (BGIS) identifies a loss-of-function mutant of the DEK oncogene, indicating DNA modulating activities of DEK in chromatin.FEBS Lett. 2021 May;595(10):1438-1453. doi: 10.1002/1873-3468.14070. Epub 2021 Mar 24. FEBS Lett. 2021. PMID: 33686684
-
High-affinity interaction of poly(ADP-ribose) and the human DEK oncoprotein depends upon chain length.Biochemistry. 2010 Aug 24;49(33):7119-30. doi: 10.1021/bi1004365. Biochemistry. 2010. PMID: 20669926 Free PMC article.
-
Control of tumorigenesis and chemoresistance by the DEK oncogene.Clin Cancer Res. 2010 Jun 1;16(11):2932-8. doi: 10.1158/1078-0432.CCR-09-2330. Epub 2010 May 25. Clin Cancer Res. 2010. PMID: 20501624 Free PMC article. Review.
-
The DEK protein--an abundant and ubiquitous constituent of mammalian chromatin.Gene. 2004 Dec 8;343(1):1-9. doi: 10.1016/j.gene.2004.08.029. Gene. 2004. PMID: 15563827 Review.
Cited by
-
Intercellular trafficking of the nuclear oncoprotein DEK.Proc Natl Acad Sci U S A. 2013 Apr 23;110(17):6847-52. doi: 10.1073/pnas.1220751110. Epub 2013 Apr 8. Proc Natl Acad Sci U S A. 2013. PMID: 23569252 Free PMC article.
-
Doxorubicin induces prolonged DNA damage signal in cells overexpressing DEK isoform-2.PLoS One. 2022 Oct 3;17(10):e0275476. doi: 10.1371/journal.pone.0275476. eCollection 2022. PLoS One. 2022. PMID: 36190960 Free PMC article.
-
Overexpression of the cellular DEK protein promotes epithelial transformation in vitro and in vivo.Cancer Res. 2009 Mar 1;69(5):1792-9. doi: 10.1158/0008-5472.CAN-08-2304. Epub 2009 Feb 17. Cancer Res. 2009. PMID: 19223548 Free PMC article.
-
DEK associates with tumor stage and outcome in HPV16 positive oropharyngeal squamous cell carcinoma.Oncotarget. 2017 Apr 4;8(14):23414-23426. doi: 10.18632/oncotarget.15582. Oncotarget. 2017. PMID: 28423581 Free PMC article.
-
IRAK1 is a novel DEK transcriptional target and is essential for head and neck cancer cell survival.Oncotarget. 2015 Dec 22;6(41):43395-407. doi: 10.18632/oncotarget.6028. Oncotarget. 2015. PMID: 26527316 Free PMC article.
References
-
- Alexiadis V, Halmer L, Gruss C. Influence of core histone acetylation on SV40 minichromosomes replication in vitro. Chromosoma. 1997;105:324–331. - PubMed
-
- Ausio J, Dong F, van Holde KE. Use of selectively trysinized nucleosome core particles to analyze the role of the histone “tails” in the stabilization of the nucleosome. J Mol Biol. 1989;206:451–463. - PubMed
-
- Cairns BR, Lorch Y, Li Y, Zhang M, Lacomis L, Erdjument-Bromage H, Tempst P, Du J, Laurent B, Kornberg RD. RSC, an essential, abundant chromatin-remodeling complex. Cell. 1996;87:1249–1260. - PubMed
-
- Chang L, Loranger SS, Mizzen C, Ernst SG, Allis CD, Annunziato AT. Histones in transit: Cytosolic histone complexes and diacetylation of H4 during nucleosome assembly in human cells. Biochemistry. 1997;36:469–480. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
